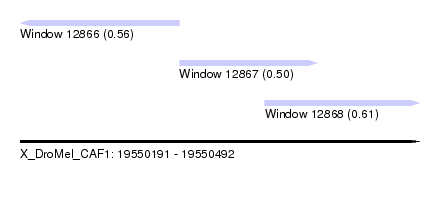
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,550,191 – 19,550,492 |
| Length | 301 |
| Max. P | 0.606562 |

| Location | 19,550,191 – 19,550,311 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19550191 120 - 22224390 GCGGGGAGCGUUUAACAACGAACGUUGGCGUUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUUGCUAUUUUUAAAAGCGGCGUAUUAAGAACCGUAGGCCAGCGA (((..((((((....((((....))))))))))..))).......((((.(((((....))..............((((((.........))))))...........))).))))..... ( -36.80) >DroSec_CAF1 13736 116 - 1 GCGGGGAGCGUUUAA---CGAACGUUGGCGUUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUUGCUAUUUUUA-AAGCGGCGUAUUAAGAGCCGUAGGCCAGCGA (((..(((((((.((---(....))))))))))..))).......((((((((((..((((.......))))...)))))).......-..(((((.........))))).))))..... ( -40.80) >DroSim_CAF1 14675 116 - 1 GCGGGGAGCGUUUAA---CGAACGUUGGCGUUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUUGCUAUUUUUA-AAGCGGCGUAUUAAGAGCCGUAGGCCAGCGA (((..(((((((.((---(....))))))))))..))).......((((((((((..((((.......))))...)))))).......-..(((((.........))))).))))..... ( -40.80) >DroEre_CAF1 13692 110 - 1 GCGGGGAGCGUUUAA---CGAACG------UUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUAGCUAUUUUUA-AAGCGGCGUAUUAAGAACCGUAGGCCAGCGA (((..((((((((..---.)))))------)))..))).......((((.(((((....))..............((.(((.......-.))).))...........))).))))..... ( -32.80) >DroYak_CAF1 13402 110 - 1 GCGGGGAGCGUUUAA---CGAACG------UUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUUGCUAUUUUUA-AAGCGGCGUAUUAAGAACCGUAGGCCAGCGA (((..((((((((..---.)))))------)))..))).......((((.(((((....))..............((((((.......-.))))))...........))).))))..... ( -35.80) >consensus GCGGGGAGCGUUUAA___CGAACGUUGGCGUUUUUCGCCAAUGACGGCCAGCGACGAGAGUACAUAAGAUUUAUAGUUGCUAUUUUUA_AAGCGGCGUAUUAAGAACCGUAGGCCAGCGA (((..((((((((......)))))......)))..))).......((((.(((((....))..............((((((.........))))))...........))).))))..... (-28.12 = -28.32 + 0.20)


| Location | 19,550,311 – 19,550,415 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.84 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -29.02 |
| Energy contribution | -29.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19550311 104 + 22224390 UGA--------UCUCGAUAUAUUGG--------GAUCUUGGUGCCGCCGGUGCUGUGGCAGCGUGUAAAGUAAAGGCAUCGCAUCCCCGAGCCCAUGUAAAUCGGCCACAUGCACUCACU .((--------((((((....))))--------))))..((((.....(((((((((((.(((((.........(((.(((......)))))))))))......)))))).))))))))) ( -34.30) >DroSec_CAF1 13852 102 + 1 UGA--------UCUCGAGAUCUUGG--------GAUCUUGGUGCCGCCGGUGCUGUGGCAGCGUGUAAAGUAAGAGCAUCGCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACU .((--------((((((....))))--------))))..((((.....(((((((((((.((((((.........))).)))...--(((...........))))))))).))))))))) ( -34.60) >DroSim_CAF1 14791 110 + 1 UGA--------UCUCGAGAUCUUGGGAUCUUGGGAUCUUGGUGCCGCCGGUGCUGUGGCAGCGUGUAAAGUAAGAGCAUCGCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACU .((--------((((((((((....))))))))))))..((((.....(((((((((((.((((((.........))).)))...--(((...........))))))))).))))))))) ( -43.20) >DroEre_CAF1 13802 110 + 1 UGAUUUCGAGAUCUCGAGAUCUUGG--------GAUCUUGGUGCCGCCGGUGCUGUGGCAGUGUGUAAAGUAAGAGCAUCACAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACU .(((..(((((((....))))))).--------.)))..((((.....(((((((((((.((((((.........))).)))...--(((...........))))))))).))))))))) ( -39.10) >DroYak_CAF1 13512 102 + 1 UGA--------UCUCGAGAUCUUGG--------GAUCUUGGUGCCGCCGGUGCUGUGGCAGUGUGUAAAGUAAAAGCAUCGCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACU .((--------((((((....))))--------))))..((((.....(((((((((((.((((((.........))).)))...--(((...........))))))))).))))))))) ( -32.70) >consensus UGA________UCUCGAGAUCUUGG________GAUCUUGGUGCCGCCGGUGCUGUGGCAGCGUGUAAAGUAAGAGCAUCGCAUC__CGAGCCCAUGUAAAUCGGCCACAUGCACUCACU ..............(((((((............)))))))(((.....(((((((((((.((((((.........))).))).....(((...........))))))))).)))))))). (-29.02 = -29.02 + -0.00)



| Location | 19,550,375 – 19,550,492 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -22.82 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19550375 117 + 22224390 GCAUCCCCGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGUCAGUCC---AUGGCACACACUUAACAAAAUCGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA (((....(((.....(((...(((((....((....))....))))).....(((((....---.)))))........)))...)))...)))((((((((.........)))))))).. ( -24.20) >DroSec_CAF1 13916 115 + 1 GCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGUCAGUCC---AUGGCACACACUUAACAAAAUCGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA (((..--(((.....(((...(((((....((....))....))))).....(((((....---.)))))........)))...)))...)))((((((((.........)))))))).. ( -24.60) >DroSim_CAF1 14863 118 + 1 GCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGUCAGUCCACCAUGGCACACACUUAACAAAAUCGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA (((..--(((.....(((...(((((....((....))....))))).....(((((........)))))........)))...)))...)))((((((((.........)))))))).. ( -24.60) >DroEre_CAF1 13874 115 + 1 ACAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGCCAGUCC---AUGGCACACACUUAACAAAAUCGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA .....--(((.....(((...(((((....((....))....))))).....(((((....---.)))))........)))...)))......((((((((.........)))))))).. ( -25.40) >DroYak_CAF1 13576 115 + 1 GCAUC--CGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGCCUGUCC---AUGGCACACACUUAACAAAAACGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA (((..--((.....((((...(((((....((....))....))))))))).((((.....---..))))...............))...)))((((((((.........)))))))).. ( -24.30) >consensus GCAUC__CGAGCCCAUGUAAAUCGGCCACAUGCACUCACUCAGCCGAACAUCUGUCAGUCC___AUGGCACACACUUAACAAAAUCGACCUGCCACGUCCUCAAUCGAACAGGAUGUGAA (((....(((.....(((...(((((....((....))....))))).....(((((........)))))........)))...)))...)))((((((((.........)))))))).. (-22.82 = -23.18 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:56 2006