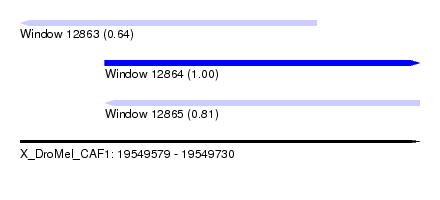
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,549,579 – 19,549,730 |
| Length | 151 |
| Max. P | 0.999483 |

| Location | 19,549,579 – 19,549,691 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.35 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -25.12 |
| Energy contribution | -25.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19549579 112 - 22224390 UAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGGACGAGUGUCUGUCGACAAAUUCGAAUAGCGUU-------- .......(((((..(((.(((((......(((((...(((((....))))))))))))))).))).....((((((.(((((....)))))))))))..)))))........-------- ( -34.40) >DroSec_CAF1 13185 116 - 1 UAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGGACGAGUGUCUGUGGACAAACUCGAAUAGCGAUC----GAU ......(((((...((......((((((..((((((.(((((....))))).((((......)))).))))))..))))))((((((((....)))..)))))....))..))----))) ( -34.40) >DroSim_CAF1 14123 120 - 1 UAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGGACGAGUGUCUGUGGCCAAACUCGAAUAGCGAUCGAUCGAU ......(((((((.(((.(((((......(((((...(((((....))))))))))))))).))).))....(((((((..(((((((.....))...))))).....)))))))))))) ( -34.10) >DroEre_CAF1 13105 112 - 1 CAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUUCUAAGCACAUGCAGAUAUAUGCACUACUAUCGAUCGGGCGAGUGUAUAUGGGCAGAAUCGAAUAGCCAU-------- ......(((((((........))))))).(((((...(((((....))))))))))..((((((((((....((....))...))))))))))(((...........)))..-------- ( -27.10) >DroYak_CAF1 12780 112 - 1 UAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUACUUCUAAGCACAUGCAGAUAUAUGCACUACUGCCGAUCGGACGAGUGUCUAUGGGCAAAAUAGAAUAACCAU-------- ..............(((.((((((.((..(((.....)))..))...((....)))))))).))).(((.((((.((.((((....)))))).))))...))).........-------- ( -20.70) >consensus UAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGGACGAGUGUCUGUGGACAAAAUCGAAUAGCGAU________ ..(((.(((((((.(((.(((((......(((((...(((((....))))))))))))))).))).))....))))).)))(((.((((....))))...)))................. (-25.12 = -25.80 + 0.68)



| Location | 19,549,611 – 19,549,730 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -33.52 |
| Energy contribution | -33.24 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19549611 119 + 22224390 CCGAUCGACAUUAGUGCAUAUAUCUGCAUGUGCUUAGCAGCACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUAAAAUCGGUCGGGGCUUU-GCGCAAGCCAAGAUCGAAGAUA (((.((((....((.(((.((((.((((((((((....)))))...))))).......)))).))).)))))).)))......((((((..(((((.-....)))))..))))))..... ( -34.80) >DroSec_CAF1 13221 120 + 1 CCGAUCGACAUUAGUGCAUAUAUCUGCAUGUGCUUAGCAGCACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUAAAAGCGGUCGGGGCUUUAGCGCAAGCCAAGAUCGAAGAUU .((((((.((((.(((((......)))))(((((....)))))..)))))..............((((..((((((........))))))))))....((....))...)))))...... ( -33.80) >DroSim_CAF1 14163 120 + 1 CCGAUCGACAUUAGUGCAUAUAUCUGCAUGUGCUUAGCAGCACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUAAAAUCGGUCGGGGCUUUAGCGCAAGCCAAGAUCGAAGAUU (((.((((....((.(((.((((.((((((((((....)))))...))))).......)))).))).)))))).)))......((((((..(((((......)))))..))))))..... ( -35.00) >DroEre_CAF1 13137 119 + 1 CCGAUCGAUAGUAGUGCAUAUAUCUGCAUGUGCUUAGAAGCACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUGAAAUCGGUCGGGGCUUU-GCGCAAGUCUAGAUCGAAAAUU .(((((((((((((((((......)))))(((((....)))))....))).)))).....((((((.((((((((((......))))))))))....-..))))))...)))))...... ( -35.70) >DroYak_CAF1 12812 119 + 1 CCGAUCGGCAGUAGUGCAUAUAUCUGCAUGUGCUUAGAAGUACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUAAAAUCGGUCGGGGCUUU-GCGCAAGCCAAGAUCGAAGAUU .((((((((....(((((......((((((((((....)))))...))))).............((((..(((((((......)))))))))))..)-))))..)))..)))))...... ( -35.20) >consensus CCGAUCGACAUUAGUGCAUAUAUCUGCAUGUGCUUAGCAGCACUCAAUGCAUAUCAAAAUAUUUGCUCUUCGACUGGAUAAAAUCGGUCGGGGCUUU_GCGCAAGCCAAGAUCGAAGAUU (((.((((....((.(((.((((.((((((((((....)))))...))))).......)))).))).)))))).)))......((((((..(((((......)))))..))))))..... (-33.52 = -33.24 + -0.28)


| Location | 19,549,611 – 19,549,730 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.49 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19549611 119 - 22224390 UAUCUUCGAUCUUGGCUUGCGC-AAAGCCCCGACCGAUUUUAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGG ......(((((..(((((....-.)))))..((((((((......))))).((.(((.(((((......(((((...(((((....))))))))))))))).))).))...)))))))). ( -34.60) >DroSec_CAF1 13221 120 - 1 AAUCUUCGAUCUUGGCUUGCGCUAAAGCCCCGACCGCUUUUAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGG ......(((((..(((((......)))))......((...((((..(((((((........))))))).(((((...(((((....))))))))))))))...)).........))))). ( -32.30) >DroSim_CAF1 14163 120 - 1 AAUCUUCGAUCUUGGCUUGCGCUAAAGCCCCGACCGAUUUUAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGG ......(((((..(((((......)))))..((((((((......))))).((.(((.(((((......(((((...(((((....))))))))))))))).))).))...)))))))). ( -34.80) >DroEre_CAF1 13137 119 - 1 AAUUUUCGAUCUAGACUUGCGC-AAAGCCCCGACCGAUUUCAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUUCUAAGCACAUGCAGAUAUAUGCACUACUAUCGAUCGG ......((((((((.(((((..-...))..((((.(........).))))))).(((.(((((......(((((...(((((....))))))))))))))).)))....)))..))))). ( -26.70) >DroYak_CAF1 12812 119 - 1 AAUCUUCGAUCUUGGCUUGCGC-AAAGCCCCGACCGAUUUUAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUACUUCUAAGCACAUGCAGAUAUAUGCACUACUGCCGAUCGG ......(((((..(((((....-.))))).((((.(........).)))).((.(((.((((((.((..(((.....)))..))...((....)))))))).))).))......))))). ( -26.20) >consensus AAUCUUCGAUCUUGGCUUGCGC_AAAGCCCCGACCGAUUUUAUCCAGUCGAAGAGCAAAUAUUUUGAUAUGCAUUGAGUGCUGCUAAGCACAUGCAGAUAUAUGCACUAAUGUCGAUCGG ......(((((..(((((......))))).((((.(........).)))).((.(((.(((((......(((((...(((((....))))))))))))))).))).))......))))). (-29.20 = -29.60 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:53 2006