
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,532,427 – 19,532,556 |
| Length | 129 |
| Max. P | 0.872300 |

| Location | 19,532,427 – 19,532,539 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.62 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
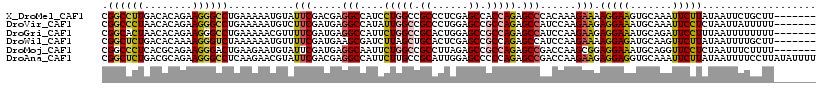
>X_DroMel_CAF1 19532427 112 + 22224390 CGGCCUUGACACAGAAGGGCCUGAAAAAUGUAUUCGACGAGGCCAUCCUGGCCGCCCUCGAGCCACCAGAGCCCACAAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUGCUU------- ...........((((((((((((......((.(((((.(.((((.....))))..).)))))..))))).)))).........(((((.......)))))....)))))...------- ( -28.60) >DroVir_CAF1 577 112 + 1 CGGCCCUAACACAGAAGGGCCUGAAAAAUGUCUUCGAUGAGGCCAUAUUGGCCGCCCUGGAGCCGCCAGAGCCAUCCAAGAAGAGGAAAUGCAAAUUCCUCUAAUUAUUUUU------- .((((((........))))))..(((((((((((.((((.((((.....))))((.((((.....)))).)))))).))))(((((((.......)))))))...)))))))------- ( -43.90) >DroGri_CAF1 568 112 + 1 CGGCACUAACACAGAAGGGCCUGAAAAACGUUUUCGAUGAGGCCAUUCUGGCCGCACUGGAGCCGCCAGAGCCAUCCAAGAAGAGGAAAUGCAGAUUCCUUUAAUUUUUUUU------- .(((.............(((((((((.....))))....)))))..((((((.((......)).)))))))))........(((((((.......)))))))..........------- ( -33.00) >DroWil_CAF1 4883 112 + 1 CGGCUCUGACACAAAAGGGUCUAAAAAAUGUUUUCGAUGAAGCGAUCUUAGCUGCACUCGAGCCGCCAGAGCCAUCCAAGAAAAGGAGAUGCAAGUUCUUAUAAUUUUGCUU------- .(((((((.......((((((........(((((....))))))))))).((.((......)).))))))))).(((.......)))...(((((..........)))))..------- ( -25.90) >DroMoj_CAF1 576 112 + 1 CGGCCCUCACGCAGAAGGGACUGAAGAAUGUAUUCGAUGAGGCAAUUCUGGCCGCCUUAGAGCCGCCAGAGCCGACCAAGCGGAGGAAAUGCAGGUUCCUCUAAUUUCUUUU------- (((((((((....(((...((........)).)))..)))))....((((((.((......)).)))))))))).......(((((((.......)))))))..........------- ( -35.10) >DroAna_CAF1 546 119 + 1 CGGCUCUGACGCAGAAGGGCCUCAAGAACGUAUUCGACGAGGCCAUUCUUGCCGCAUUGGAGCCCCCAGAGCCGACCAAGAAGAGGAGGUGCAAAUUCUUAUAAUUUUCCUUAUAUUUU ((((((((..((((((.((((((..(((....)))...)))))).))).))).((......))...))))))))........(((((((................)))))))....... ( -39.29) >consensus CGGCCCUGACACAGAAGGGCCUGAAAAAUGUAUUCGAUGAGGCCAUUCUGGCCGCACUGGAGCCGCCAGAGCCAACCAAGAAGAGGAAAUGCAAAUUCCUAUAAUUUUUCUU_______ .((((((........))))))...........(((.....(((....(((((.((......)).))))).)))......))).(((((.......)))))................... (-20.53 = -20.62 + 0.09)


| Location | 19,532,427 – 19,532,539 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19532427 112 - 22224390 -------AAGCAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUUGUGGGCUCUGGUGGCUCGAGGGCGGCCAGGAUGGCCUCGUCGAAUACAUUUUUCAGGCCCUUCUGUGUCAAGGCCG -------..(((((.((....)).))))).................(((..((..(.(..((((((((((.....)))).....(((.......)))..))))))..))..))..))). ( -33.50) >DroVir_CAF1 577 112 - 1 -------AAAAAUAAUUAGAGGAAUUUGCAUUUCCUCUUCUUGGAUGGCUCUGGCGGCUCCAGGGCGGCCAAUAUGGCCUCAUCGAAGACAUUUUUCAGGCCCUUCUGUGUUAGGGCCG -------((((((....(((((((.......)))))))((((.(((((((((((.....)))))))((((.....)))).)))).)))).))))))..((((((........)))))). ( -48.70) >DroGri_CAF1 568 112 - 1 -------AAAAAAAAUUAAAGGAAUCUGCAUUUCCUCUUCUUGGAUGGCUCUGGCGGCUCCAGUGCGGCCAGAAUGGCCUCAUCGAAAACGUUUUUCAGGCCCUUCUGUGUUAGUGCCG -------............(((((.......)))))..........(((.(((((((((.......))))((((.(((((....((((.....))))))))).))))..))))).))). ( -32.40) >DroWil_CAF1 4883 112 - 1 -------AAGCAAAAUUAUAAGAACUUGCAUCUCCUUUUCUUGGAUGGCUCUGGCGGCUCGAGUGCAGCUAAGAUCGCUUCAUCGAAAACAUUUUUUAGACCCUUUUGUGUCAGAGCCG -------..((((............))))...(((.......))).(((((((((...((((.((.(((.......))).))))))..(((...............)))))))))))). ( -28.16) >DroMoj_CAF1 576 112 - 1 -------AAAAGAAAUUAGAGGAACCUGCAUUUCCUCCGCUUGGUCGGCUCUGGCGGCUCUAAGGCGGCCAGAAUUGCCUCAUCGAAUACAUUCUUCAGUCCCUUCUGCGUGAGGGCCG -------..(((((....((((((.......))))))...(((((.(((((((((.(((....))).))))))...)))..))))).....)))))..(.(((((......))))).). ( -35.80) >DroAna_CAF1 546 119 - 1 AAAAUAUAAGGAAAAUUAUAAGAAUUUGCACCUCCUCUUCUUGGUCGGCUCUGGGGGCUCCAAUGCGGCAAGAAUGGCCUCGUCGAAUACGUUCUUGAGGCCCUUCUGCGUCAGAGCCG ........(((.(((((.....)))))...)))((.......)).(((((((((..((......)).(((.(((.(((((((..(((....))).))))))).)))))).))))))))) ( -38.30) >consensus _______AAAAAAAAUUAUAAGAAUUUGCAUUUCCUCUUCUUGGAUGGCUCUGGCGGCUCCAGGGCGGCCAGAAUGGCCUCAUCGAAUACAUUUUUCAGGCCCUUCUGUGUCAGGGCCG ..............................................((((((((((((......))...(((((.(((((....(((....)))...))))).))))))))))))))). (-22.49 = -22.80 + 0.31)



| Location | 19,532,466 – 19,532,556 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -20.79 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19532466 90 + 22224390 AGGCCAUCCUGGCCGCCCUCGAGCCACCAGAGCCCACAAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUGCUU--UCC--UCACACGCGGUAUC .((((.....))))((((((.........)))............((((((.(((((((.....)))).)))))--)))--).......)))... ( -19.30) >DroVir_CAF1 616 89 + 1 AGGCCAUAUUGGCCGCCCUGGAGCCGCCAGAGCCAUCCAAGAAGAGGAAAUGCAAAUUCCUCUAAUUAUUUUU--GUUU-C--UUUGUUGUUUU .((((.....))))((.((((.....)))).)).........(((((((.......)))))))..........--....-.--........... ( -23.60) >DroPse_CAF1 601 89 + 1 AGGCCAUACUGGCCGCCCUGGAGCCACCAGAACCAACCAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUUUUUUUUUUC-UUUCUC----UCUC .((((.....))))...((((.....))))........((((((((((((...................))))))))))-))....----.... ( -18.31) >DroEre_CAF1 572 90 + 1 AGGCCAUCCUGGCCGCUCUGGAGCCACCAGAGCCCACAAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUUUUU--UUC--CCACACGCGGUAUC .((((.....))))(((((((.....)))))))......((((((((((................))))))))--)).--((......)).... ( -24.99) >DroYak_CAF1 572 92 + 1 AGGCCAUUCUGGCCGCCCUGGAGCCACCAGAGCCCACAAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUUCUU--UCCCCCCACACGCGGUAUC .((((.....))))(((((((.....)))).((..........(((((((.................))))))--)..........)))))... ( -20.08) >DroPer_CAF1 3044 87 + 1 AGGCCAUACUGGCCGCCCUGGAGCCACCAGAACCAACCAAGAAAAGGAAGUGCAAAUUCUUAUAAUU--UUUUUUUUUC-UUUCUC----UCUC .((((.....))))...((((.....))))........((((((((((((.................--))))))))))-))....----.... ( -18.43) >consensus AGGCCAUACUGGCCGCCCUGGAGCCACCAGAGCCAACAAAGAAAAGGAAGUGCAAAUUCUUAUAAUUCUUUUU__UUCC_CCACACGCGGUAUC .((((.....))))((.((((.....)))).))...........(((((.......)))))................................. (-14.59 = -14.95 + 0.36)



| Location | 19,532,466 – 19,532,556 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.95 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
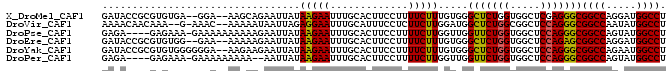
>X_DroMel_CAF1 19532466 90 - 22224390 GAUACCGCGUGUGA--GGA--AAGCAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUUGUGGGCUCUGGUGGCUCGAGGGCGGCCAGGAUGGCCU ....(((((...((--(((--(.(((((.((....)).)))))..)))))).....)))))(((((.(.....).)))))((((.....)))). ( -31.40) >DroVir_CAF1 616 89 - 1 AAAACAACAAA--G-AAAC--AAAAAUAAUUAGAGGAAUUUGCAUUUCCUCUUCUUGGAUGGCUCUGGCGGCUCCAGGGCGGCCAAUAUGGCCU ....((.(.((--(-(...--..........(((((((.......))))))))))).).))(((((((.....)))))))((((.....)))). ( -28.11) >DroPse_CAF1 601 89 - 1 GAGA----GAGAAA-GAAAAAAAAAAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUGGUUGGUUCUGGUGGCUCCAGGGCGGCCAGUAUGGCCU ..((----(((((.-(((........(((((.....)))))....))).)))))))(((((...(((((.(((....))).)))))..))))). ( -24.10) >DroEre_CAF1 572 90 - 1 GAUACCGCGUGUGG--GAA--AAAAAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUUGUGGGCUCUGGUGGCUCCAGAGCGGCCAGGAUGGCCU ....(((((...((--(((--.....(((((.....)))))....)))))......)))))(((((((.....)))))))((((.....)))). ( -31.90) >DroYak_CAF1 572 92 - 1 GAUACCGCGUGUGGGGGGA--AAGAAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUUGUGGGCUCUGGUGGCUCCAGGGCGGCCAGAAUGGCCU ....(((((...(((((((--(....(((((.....)))))....))))))))...)))))(((((((.....)))))))((((.....)))). ( -34.30) >DroPer_CAF1 3044 87 - 1 GAGA----GAGAAA-GAAAAAAAAA--AAUUAUAAGAAUUUGCACUUCCUUUUCUUGGUUGGUUCUGGUGGCUCCAGGGCGGCCAGUAUGGCCU ..((----(((((.-(((......(--((((.....)))))....))).)))))))(((((...(((((.(((....))).)))))..))))). ( -24.90) >consensus GAUACCGCGAGUGA_GAAA__AAAAAGAAUUAUAAGAAUUUGCACUUCCUUUUCUUGGUGGGCUCUGGUGGCUCCAGGGCGGCCAGUAUGGCCU .................................(((((.............))))).....(((((((.....)))))))((((.....)))). (-18.95 = -18.62 + -0.33)
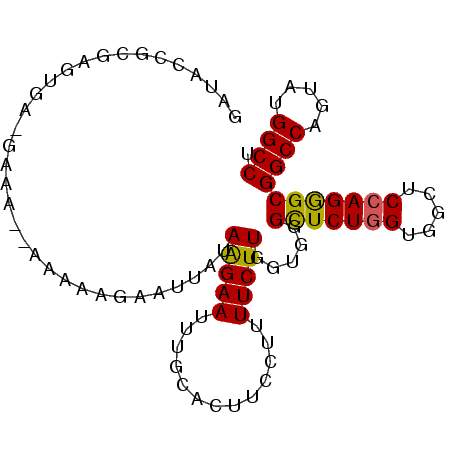

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:43 2006