
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,483,925 – 19,484,085 |
| Length | 160 |
| Max. P | 0.965585 |

| Location | 19,483,925 – 19,484,045 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -41.32 |
| Consensus MFE | -39.98 |
| Energy contribution | -39.26 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.97 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19483925 120 + 22224390 UAUCGUGUUCUAGUCGAAUAUAUGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGCGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ..(((((..((.((((.((...(((((((((.....((((.(....).)))).)))))))))..)).))))))...)))))(((..(((...(((.....)))..)))..)))....... ( -48.20) >DroSec_CAF1 5910 120 + 1 UAUCGUAUUCUAGUCGAAUAUAGGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ..(((((((((...(.......)((((((........((((((......)))))))))))))))))))))..(((..(((.(((..(((...(((.....)))..)))..))).)))))) ( -39.60) >DroSim_CAF1 6860 120 + 1 UAUCGUAUUCUAGUCGAAUAUAGGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ..(((((((((...(.......)((((((........((((((......)))))))))))))))))))))..(((..(((.(((..(((...(((.....)))..)))..))).)))))) ( -39.60) >DroEre_CAF1 5591 120 + 1 UAUCGUAUUCUAAUCGGAUAUAGGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGUGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ..((((..((((..((((........))))))))(((((((.((..(..(((......)))..)...)).)))))))))))(((..(((...(((.....)))..)))..)))....... ( -38.80) >DroYak_CAF1 5146 120 + 1 UAUCGUAUUCUAGUCGGAUAUAGGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ..((((..((((..((((........))))))))(((((((.((..(..(((......)))..)...)).)))))))))))(((..(((...(((.....)))..)))..)))....... ( -40.40) >consensus UAUCGUAUUCUAGUCGAAUAUAGGCCUCCGUGGAGAGGCUGGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGU ((((((((((.....))))))..((((((........((((((......))))))))))))..)))).....(((..(((.(((..(((...(((.....)))..)))..))).)))))) (-39.98 = -39.26 + -0.72)

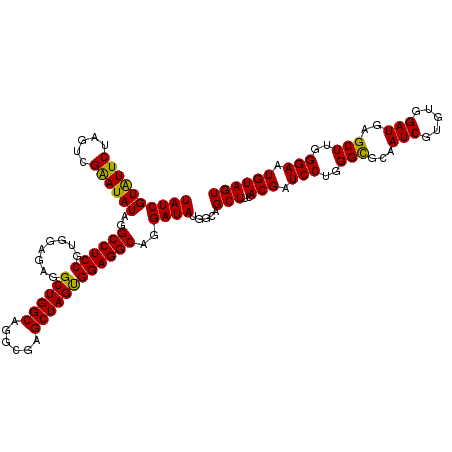
| Location | 19,483,965 – 19,484,085 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -41.86 |
| Energy contribution | -41.22 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19483965 120 + 22224390 GGCAGGCGAGCUAGCGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUGGAGGAGGGCUCAUGCUUAAG ((((...(((((..(((((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))))......))))).)))).... ( -43.70) >DroSec_CAF1 5950 120 + 1 GGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUAGAGGAGGGCUCAUGCUUAAG ((((...(((((..(((((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))))......))))).)))).... ( -42.60) >DroSim_CAF1 6900 120 + 1 GGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUGGAGCAGGGCUCAUGCUUAAG ((((...(((((.((.(((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))....))..))))).)))).... ( -42.20) >DroEre_CAF1 5631 120 + 1 GGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGUGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUGGAGCAGGGCUCAUGUUUGAG ..((((((((((.((.(((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))....))..)))))..))))).. ( -39.90) >DroYak_CAF1 5186 120 + 1 GGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUGGAGCAGGGCUCAUGCUUGAG ..((((((((((.((.(((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))....))..)))))..))))).. ( -44.00) >consensus GGCAGGCGAGCUAGUGGAGGCAGGAUAUGGCAGCUUCACGAUCCUGGGCGCAAUCGUGUGGAUGAGCUUGGGAAUGUAGUUGACCAUGGCGAAGUCUUUGGAGCAGGGCUCAUGCUUAAG ((((...(((((..(((((((....((((((((((..(((.(((..(((...(((.....)))..)))..))).)))))))).))))).....)))))))......))))).)))).... (-41.86 = -41.22 + -0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:22 2006