
| Sequence ID | X_DroMel_CAF1 |
|---|---|
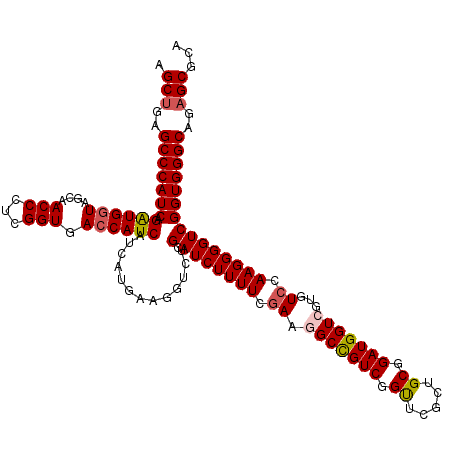
| Location | 19,482,618 – 19,482,861 |
| Length | 243 |
| Max. P | 0.958417 |

| Location | 19,482,618 – 19,482,738 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -48.12 |
| Consensus MFE | -43.84 |
| Energy contribution | -44.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19482618 120 + 22224390 AGCUGAGCCCAUCAGGUGGUAGCAACCCUCGGUGACCAUCAUCAUGAAGGUCUGCAUCUUUUCGAAGGCCGUCGGUUCGCUGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCA .(((..(((((((.....(((((((((..((((((((.((.....)).)))).....(((....)))))))..)))).)))))((((......))))........)))))))..)))... ( -48.40) >DroSec_CAF1 4601 120 + 1 AGCUGAGCCCAUCAGAUGGUAGCAACCCUCGGUGACCAUCAUCAUGAAGGUCUGCAUCUUUUCGAAGGCCGUCGGUUCGCUGCGGAUGGUCGCGUCCAAGGGGUCGGUGGGCAGAGCGCA .(((..(((((((.(((.(((((((((..((((((((.((.....)).)))).....(((....)))))))..)))).)))))(((((....))))).....))))))))))..)))... ( -51.30) >DroSim_CAF1 4665 120 + 1 AGCUGAGCCCAUCAGAUGGUAGCAACCCUCGGUGACCAUCAUCAUGAAGGUCUGCAUCUUUUCGAAGGCCGUCGGUUCGCUGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCA .(((..(((((((.(((.(((((((((..((((((((.((.....)).)))).....(((....)))))))..)))).)))))((((......)))).....))))))))))..)))... ( -49.40) >DroEre_CAF1 4284 120 + 1 UGCUGAGCCCAUCAGAUGGUAGCAACCCUCGGUGACCAUCAUCAUGAAUGUCUGCAUCUUUUCGAAGGCCGUCGGCUCGCUGCGGAUGGUGGUGUCCAAGGGGUCGGUGGGCAGAGCGCA .(((..(((((((.((((....)..((((.((..(((((((((......((((.(........).))))((.(((....))))))))))))))..)).))))))))))))))..)))... ( -46.70) >DroYak_CAF1 3839 120 + 1 UGCUGAGCCCAUCAGAUGAUAGCAACCCUCGGUGACCAUCAUCAUGAAUGUCUGCAUCUUUUCAAAGGCUGUCGGCUCGCUGCGGAUGGUGGUGUCCAAGGGGUCGGUGGGCAGUGCGCA .((...(((((((.((((....)..((((.((..(((((((((..(...(((.(((((((....)))).))).)))...)....)))))))))..)).))))))))))))))...))... ( -44.80) >consensus AGCUGAGCCCAUCAGAUGGUAGCAACCCUCGGUGACCAUCAUCAUGAAGGUCUGCAUCUUUUCGAAGGCCGUCGGUUCGCUGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCA .(((..(((((((.((((((....(((...))).)))))).............(.(((((((.((.(((((((.((.....)).)))))))...)).)))))))))))))))..)))... (-43.84 = -44.28 + 0.44)

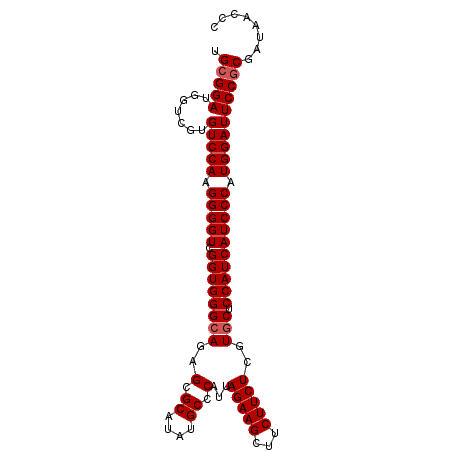
| Location | 19,482,698 – 19,482,796 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -32.61 |
| Energy contribution | -33.01 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19482698 98 + 22224390 UGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCAUAUGCCCAUUAGAAGCUUCUUCUCGUGCUCCAUCAUCCCAUGGAUUCCACGAUAACCC ...((...((((((((((.(((((.(((((((((((.((....)).)....((((...)))))).))).)))))))))).))))...))))))..)). ( -35.70) >DroSec_CAF1 4681 98 + 1 UGCGGAUGGUCGCGUCCAAGGGGUCGGUGGGCAGAGCGCAUAUGCCCAUUAGAAGCUUCUUCUCGUGCUCCAUCAUCCCAUGGAUUCCGCGAUAACCC ...((...((((((((((.(((((.(((((((((((.((....)).)....((((...)))))).))).)))))))))).))))...))))))..)). ( -37.20) >DroSim_CAF1 4745 98 + 1 UGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCAUAUGCCCAUUAGAAGCUUCUUCUCGUGCUCCAUCAUCCCAUGGAUUCCGCGAUAGCCC ...((...((((((((((.(((((.(((((((((((.((....)).)....((((...)))))).))).)))))))))).))))...))))))..)). ( -36.40) >DroEre_CAF1 4364 98 + 1 UGCGGAUGGUGGUGUCCAAGGGGUCGGUGGGCAGAGCGCAAAUGCCCAUUAGAAGCUUCUUCUCGUUCUCCAUCAUCCCAUGGAUUCCGCGAUAUCCC ...(((((((((.(((((.(((((.(((((..(((((((....)).....(((((...))))).))))))))))))))).))))).))))..))))). ( -39.90) >DroYak_CAF1 3919 98 + 1 UGCGGAUGGUGGUGUCCAAGGGGUCGGUGGGCAGUGCGCAAAUGCCCAUUAGAAGCUUCUUCUCGUGCUCCAUCAUCCCAUGGAUUCCGCGAUAUCCC ...(((((((((.(((((.(((((.(((((((((((.((....)).))))(((((...)))))...)).)))))))))).))))).))))..))))). ( -41.20) >consensus UGCGGAUGGUCGUGUCCAAGGGGUCGGUGGGCAGAGCGCAUAUGCCCAUUAGAAGCUUCUUCUCGUGCUCCAUCAUCCCAUGGAUUCCGCGAUAACCC .(((((.......(((((.(((((.((((((((..(.((....)).)...(((((...)))))..))).)))))))))).))))))))))........ (-32.61 = -33.01 + 0.40)


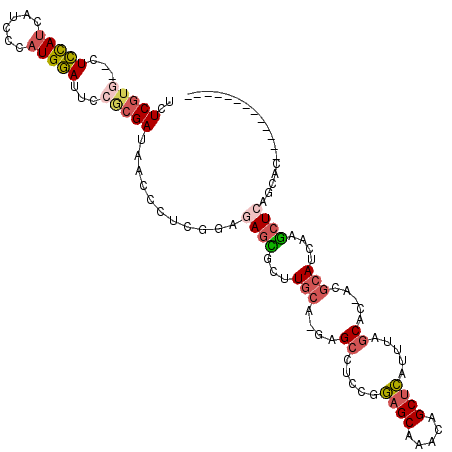
| Location | 19,482,758 – 19,482,861 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.51 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -16.33 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19482758 103 + 22224390 UCUCGUG--CUCCAUCAUCCCAUGGAUUCCACGAUAACCCUCGGAGAGCGCUUGCA-GAGCCUCCGGAGCAAACAGCUCAUUAAGCAC-ACGCAUCAAGCUCAGCAC------------- ....(((--((((((......))))).((..(((......)))..)))))).(((.-((((.....((((.....)))).....((..-..)).....)))).))).------------- ( -28.30) >DroSec_CAF1 4741 103 + 1 UCUCGUG--CUCCAUCAUCCCAUGGAUUCCGCGAUAACCCUCGGAGAGCGCGUGCA-GAGCCUCCGGAGCAAACAGCUCAUUUAGCAC-ACGCAUCAAGCUCAGCAC------------- ....(((--(((....((((...))))((((.(......).))))))))))((((.-((((.....((((.....)))).....((..-..)).....)))).))))------------- ( -33.80) >DroSim_CAF1 4805 103 + 1 UCUCGUG--CUCCAUCAUCCCAUGGAUUCCGCGAUAGCCCUCGGAGAGCGCGUGCA-GAGCCUCCGGAGCAAACAGCUCAUUUAGCAC-ACGCAUCAAGCUCAGCAC------------- ....(((--(((....((((...))))((((.(......).))))))))))((((.-((((.....((((.....)))).....((..-..)).....)))).))))------------- ( -33.80) >DroEre_CAF1 4424 103 + 1 UCUCGUU--CUCCAUCAUCCCAUGGAUUCCGCGAUAUCCCUCGGAAAGUGCUUGCA-GAGCCUCCGGAGCAAACAGCUCGUUGAGCAC-ACGCAUCAAGCUCAGCAC------------- .......--.(((((......)))))(((((.(......).))))).(((((....-((((.....((((.....)))).((((((..-..)).)))))))))))))------------- ( -28.70) >DroYak_CAF1 3979 103 + 1 UCUCGUG--CUCCAUCAUCCCAUGGAUUCCGCGAUAUCCCUCGGAGAGUGCUUGCA-AAGCCUCCGGAGCAAACAGCUCAUUGAGCAC-ACGCAUCAAGCUCAGCAC------------- .....((--((((....(((...((((........))))...)))(((.(((....-.)))))).))))))....(((..((((((..-..)).))))....)))..------------- ( -29.80) >DroPer_CAF1 8271 108 + 1 UCUCUUUAUAUGUAACAUCCCAUGUAU----CGAUAAC--------AGAAUCUGCAUCAGCUGUCGCAGCAAAUCGCAGAUUUACCACCACUAAUCAAUAUCAACAUUUCUGGUCGUCAA ..((..((((((........)))))).----.))....--------.((((((((((..((((...))))..)).))))))))((.((((..(((..........)))..)))).))... ( -19.90) >consensus UCUCGUG__CUCCAUCAUCCCAUGGAUUCCGCGAUAACCCUCGGAGAGCGCUUGCA_GAGCCUCCGGAGCAAACAGCUCAUUUAGCAC_ACGCAUCAAGCUCAGCAC_____________ ..(((((...(((((......)))))...)))))...........((((...(((....((.....((((.....)))).....)).....)))....)))).................. (-16.33 = -17.08 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:17 2006