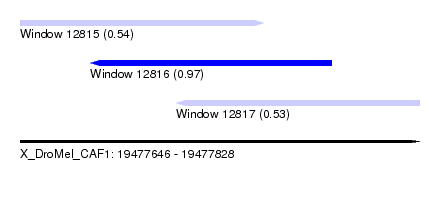
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,477,646 – 19,477,828 |
| Length | 182 |
| Max. P | 0.967737 |

| Location | 19,477,646 – 19,477,757 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.77 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -29.35 |
| Energy contribution | -30.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19477646 111 + 22224390 GUGAUAGCUGGAUAACUUGUUGGGCGAA--------UUUGUGGAAAUGGGCAACGCGAUUUCACCUGUGCCGUCGC-CAACCAUUUGCCACGUCGCAGCUGACGUUGCUGUGAAAGCGCU (((...(((.(((.....))).)))...--------...((((((((((.....(((((..((....))..)))))-...)))))).)))).(((((((.......)))))))...))). ( -37.00) >DroSec_CAF1 4555 110 + 1 GUGAUAGCUAGAUAACUAGUUGGGCGAA--------AU-GGGUAAAUGGGCAACCCGAUUUCACCUGCGACGUCGCCCAACCAUUUGCCACGUCGC-GCUGACGUUGCCGUGAAAGCGCU (((..((((((....))))))((((((.--------..-((((((((.((....)).)))).))))......))))))..........)))...((-(((.(((....)))...))))). ( -36.80) >DroEre_CAF1 4473 119 + 1 CUGAUAGCUGGAUAACUAGUUGGGCGAACCCAAGAGUUCGGUGAAAUGGGCCACCAGAUUUCACCUGUGACGUCGC-CUACCAUUUGCCACGUCACAGCUGACGUUGCCGUGAAAGCGGU ....(((((....((((..(((((....))))).)))).((((((((((....)))..))))))).(((((((.((-.........)).)))))))))))).....(((((....))))) ( -43.70) >DroYak_CAF1 6922 119 + 1 CUGAUAGCUGGAUAACCAGUUGGGCGAGACCAGCAGUUCGGUGAAAUGGGCCACCUGAUUUCACCUGUGACGUCGC-CCACCAUUUGCCACGUCACAGCUGACGUUGCCGUGAAAGCGCU ....(((((((....)))))))(((((...((((.....((((((((.((....)).))))))))((((((((.((-.........)).))))))))))))...)))))((....))... ( -47.40) >consensus CUGAUAGCUGGAUAACUAGUUGGGCGAA________UUCGGGGAAAUGGGCAACCCGAUUUCACCUGUGACGUCGC_CAACCAUUUGCCACGUCACAGCUGACGUUGCCGUGAAAGCGCU (((.(((((((....)))))))(((((............((((((((((....))..))))))))((((((((.((..........)).)))))))).......))))).......))). (-29.35 = -30.35 + 1.00)


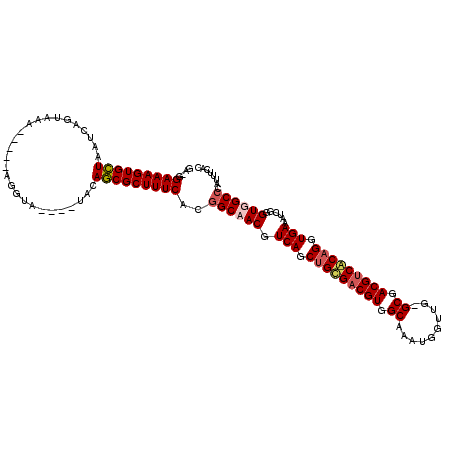
| Location | 19,477,678 – 19,477,788 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.07 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -34.46 |
| Energy contribution | -35.33 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19477678 110 - 22224390 GAAGGAAAGUGCUAAUCAGUAAA-----AGGUA----UUCAGCGCUUUCACAGCAACGUCAGCUGCGACGUGGCAAAUGGUUG-GCGACGGCACAGGUGAAAUCGCGUUGCCCAUUUCCA ...(((((((((((((((((.((-----((((.----....)).)))).)).((.(((((......))))).))...))))))-)).))((((.(.(((....))).)))))..))))). ( -37.30) >DroSec_CAF1 4586 110 - 1 GAAGGAAAGUGCUAAUCAGUAAA-----AGGUA----UACAGCGCUUUCACGGCAACGUCAGC-GCGACGUGGCAAAUGGUUGGGCGACGUCGCAGGUGAAAUCGGGUUGCCCAUUUACC ....(((((((((.(((......-----.))).----...)))))))))..((((((.(((.(-(((((((.((..(....)..)).))))))).).)))......))))))........ ( -38.90) >DroEre_CAF1 4513 114 - 1 GC-GGAAAGUGGUAAUCAGUAAUCCAUCAGGUA----UACACCGCUUUCACGGCAACGUCAGCUGUGACGUGGCAAAUGGUAG-GCGACGUCACAGGUGAAAUCUGGUGGCCCAUUUCAC ..-(..((((((...........(((((((((.----..(((((((...(((....))).)))((((((((.((.........-)).))))))))))))..))))))))).))))))..) ( -42.70) >DroYak_CAF1 6962 113 - 1 GU-GGAAAGUGCUAAUCAGUAAA-----AGGUAUCAAUCCAGCGCUUUCACGGCAACGUCAGCUGUGACGUGGCAAAUGGUGG-GCGACGUCACAGGUGAAAUCAGGUGGCCCAUUUCAC ((-((((((((((.((..(((..-----...)))..))..)))))))))..(((..(.(((.(((((((((.((.........-)).))))))))).))).....)...))).....))) ( -39.70) >consensus GA_GGAAAGUGCUAAUCAGUAAA_____AGGUA____UACAGCGCUUUCACGGCAACGUCAGCUGCGACGUGGCAAAUGGUUG_GCGACGUCACAGGUGAAAUCGGGUGGCCCAUUUCAC ....(((((((((...........................)))))))))..((((((.(((.(((((((((.((..........)).))))))))).)))......))))))........ (-34.46 = -35.33 + 0.87)


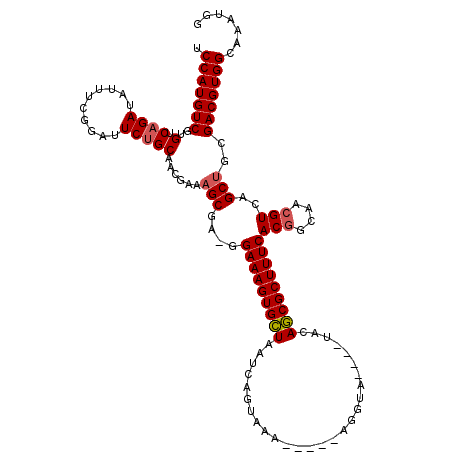
| Location | 19,477,717 – 19,477,828 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.85 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -26.08 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19477717 111 - 22224390 UCCAUGUCUUGUCAGAUAUUUGGGAUUCUGCAACGAAAGCGAAGGAAAGUGCUAAUCAGUAAA-----AGGUA----UUCAGCGCUUUCACAGCAACGUCAGCUGCGACGUGGCAAAUGG .((((((((((.((((..........))))))).(((((((...((..(((((..........-----.))))----)))..))))))).((((.......)))).)))))))....... ( -31.30) >DroSec_CAF1 4626 110 - 1 UCCAUGUCUAGUCAGAUAUUUGGGAUUCUGCAACGAAAGCGAAGGAAAGUGCUAAUCAGUAAA-----AGGUA----UACAGCGCUUUCACGGCAACGUCAGC-GCGACGUGGCAAAUGG .(((((((..(.((((..........))))).......(((...(((((((((.(((......-----.))).----...)))))))))(((....)))...)-)))))))))....... ( -32.30) >DroEre_CAF1 4552 115 - 1 UCCAUGUCGUGUCUGAUGUUUCGGAUUCUGCAAGGAAAGCGC-GGAAAGUGGUAAUCAGUAAUCCAUCAGGUA----UACACCGCUUUCACGGCAACGUCAGCUGUGACGUGGCAAAUGG .(((((((..(.((((((((.....((((....)))).((.(-((((((((((.....(((..((....))..----)))))))))))).)))))))))))).)..)))))))....... ( -40.40) >DroYak_CAF1 7001 114 - 1 UCCAUGUCGUGUCAGAUGUUUCAGAUUCUGCAAGAAGAGCGU-GGAAAGUGCUAAUCAGUAAA-----AGGUAUCAAUCCAGCGCUUUCACGGCAACGUCAGCUGUGACGUGGCAAAUGG .(((((((.((.((((..........)))))).((.((((((-(((..(((((..........-----.)))))...)))).)))))))(((((.......))))))))))))....... ( -32.30) >consensus UCCAUGUCGUGUCAGAUAUUUCGGAUUCUGCAACGAAAGCGA_GGAAAGUGCUAAUCAGUAAA_____AGGUA____UACAGCGCUUUCACGGCAACGUCAGCUGCGACGUGGCAAAUGG .(((((((..(.((((..........)))))......(((....(((((((((...........................)))))))))(((....)))..)))..)))))))....... (-26.08 = -26.46 + 0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:08 2006