
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,465,970 – 19,466,070 |
| Length | 100 |
| Max. P | 0.997179 |

| Location | 19,465,970 – 19,466,070 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -28.21 |
| Energy contribution | -29.54 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19465970 100 + 22224390 CGAUGUCCGUUACUGCGAGAACUAACGAGAUCUCGAGCCGCACCAUCACUCGUUACUGCUGCUCGGUCUCGUUAGUUUAAAAUAACAUGCAAACAGCCCC ..((((.(((....))).(((((((((((((..(((((.(((....(....)....))).))))))))))))))))))......))))((.....))... ( -30.70) >DroSec_CAF1 6348 87 + 1 CGAUGUCCGUUACUGCGAGAACUAACGAGAUCUCGAGCAGCACCAUUACUCGUUACUGCUGCUCGGUCUCGUUACAU-------------ACGUAACUUA ((((((((((....))).))..(((((((((..(((((((((..............)))))))))))))))))))))-------------.))....... ( -29.04) >DroSim_CAF1 6365 100 + 1 CGAUGUCCGUUACUGCGAGAACUAACGAGAACUCGAGCAGCACCAUUACUCGUUACUGCUGCUCGGUCUCGUUAGUUUGGAAUACCAUGCAAGCAGAUUA ............((((..((((((((((((.(.(((((((((..............))))))))))))))))))))))((....))......)))).... ( -38.94) >consensus CGAUGUCCGUUACUGCGAGAACUAACGAGAUCUCGAGCAGCACCAUUACUCGUUACUGCUGCUCGGUCUCGUUAGUUU__AAUA_CAUGCAAGCAGCUUA .......(((....))).((((((((((((.(.(((((((((..............))))))))))))))))))))))...................... (-28.21 = -29.54 + 1.33)

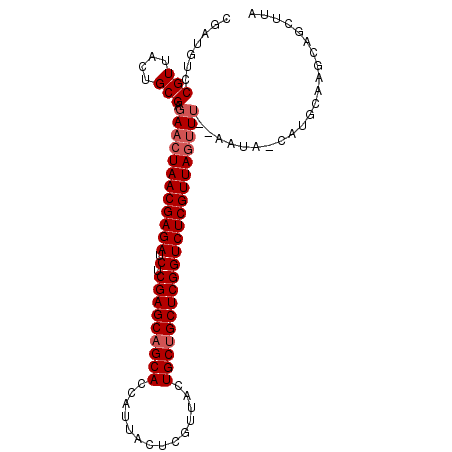
| Location | 19,465,970 – 19,466,070 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -28.57 |
| Energy contribution | -29.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19465970 100 - 22224390 GGGGCUGUUUGCAUGUUAUUUUAAACUAACGAGACCGAGCAGCAGUAACGAGUGAUGGUGCGGCUCGAGAUCUCGUUAGUUCUCGCAGUAACGGACAUCG ((..(((((.((.(((.......(((((((((((((((((.(((.((........)).))).))))).).)))))))))))...))))))))))...)). ( -34.00) >DroSec_CAF1 6348 87 - 1 UAAGUUACGU-------------AUGUAACGAGACCGAGCAGCAGUAACGAGUAAUGGUGCUGCUCGAGAUCUCGUUAGUUCUCGCAGUAACGGACAUCG ...(((((..-------------...((((((((((((((((((.((........)).))))))))).).))))))))((....)).)))))........ ( -32.90) >DroSim_CAF1 6365 100 - 1 UAAUCUGCUUGCAUGGUAUUCCAAACUAACGAGACCGAGCAGCAGUAACGAGUAAUGGUGCUGCUCGAGUUCUCGUUAGUUCUCGCAGUAACGGACAUCG ...((((.((((.(((....)))(((((((((((((((((((((.((........)).))))))))).).)))))))))))......))))))))..... ( -41.00) >consensus UAAGCUGCUUGCAUG_UAUU__AAACUAACGAGACCGAGCAGCAGUAACGAGUAAUGGUGCUGCUCGAGAUCUCGUUAGUUCUCGCAGUAACGGACAUCG .......................(((((((((((((((((((((.((........)).))))))))).).)))))))))))................... (-28.57 = -29.57 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:48:01 2006