
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,458,712 – 19,458,855 |
| Length | 143 |
| Max. P | 0.999999 |

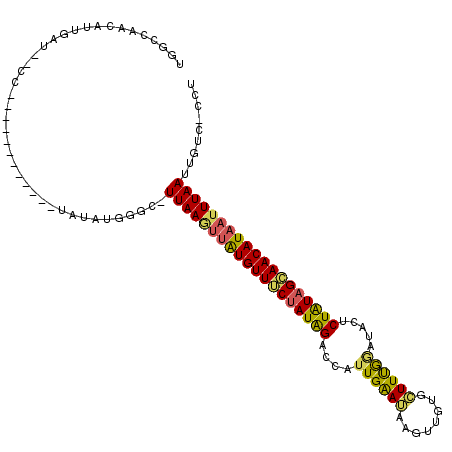
| Location | 19,458,712 – 19,458,828 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 64.78 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.67 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.57 |
| SVM decision value | 6.73 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19458712 116 + 22224390 UGGCCAACAUUGAUGUCCUACAUAUAUACAUAUAUGGGCAUUAAGUUAUGUUUCUAUGGGUCAUUGGAGAAGUUGGGCUUCGGUUACGCUGUUGAAACAUAAGUUAAUUGGUCCCA .((((((..(((((((((...((((....))))..))))))))).(((((((((.((((((.(((((((........))))))).)).)))).))))))))).....))))))... ( -35.30) >DroEre_CAF1 2562 90 + 1 UGGCC-------------------------UAUAUGAGCCUUAAGUUAUGUUUCUAUAGUCCAAUGAAUAAGUUGUGCUUUGGAUACUCUAUAGCAACAUAAUUUAAUUGUC-CAU .((((-------------------------.....).)))((((((((((((.(((((((((((...((.....))...)))))....)))))).)))))))))))).....-... ( -23.70) >DroYak_CAF1 2458 102 + 1 UGGCCAACAUUGAUAGCC------------CAUAUGGGC-UUAAAGUGUGUUGCUAUAGACUAUUGAAUAAGUUGUGAUUUAAACACUCUAUAGCAACAUCAUUUAAUUGUC-CCU .((.(((((((...((((------------(....))))-)...)))))(((((((((((...((((((........))))))....)))))))))))..........)).)-).. ( -31.90) >consensus UGGCCAACAUUGAU__CC____________UAUAUGGGC_UUAAGUUAUGUUUCUAUAGACCAUUGAAUAAGUUGUGCUUUGGAUACUCUAUAGCAACAUAAUUUAAUUGUC_CCU ........................................(((((((((((((((((((....((((((........)))))).....)))))))))))))))))))......... (-17.31 = -17.67 + 0.35)


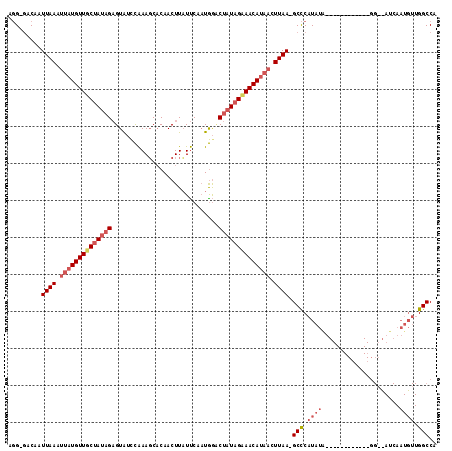
| Location | 19,458,712 – 19,458,828 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.78 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -9.16 |
| Energy contribution | -12.38 |
| Covariance contribution | 3.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19458712 116 - 22224390 UGGGACCAAUUAACUUAUGUUUCAACAGCGUAACCGAAGCCCAACUUCUCCAAUGACCCAUAGAAACAUAACUUAAUGCCCAUAUAUGUAUAUAUGUAGGACAUCAAUGUUGGCCA ..((.((((((((.(((((((((.....(((....((((.....))))....))).......))))))))).)))(((((.((((((....)))))).)).)))....))))))). ( -23.80) >DroEre_CAF1 2562 90 - 1 AUG-GACAAUUAAAUUAUGUUGCUAUAGAGUAUCCAAAGCACAACUUAUUCAUUGGACUAUAGAAACAUAACUUAAGGCUCAUAUA-------------------------GGCCA ...-.....((((.(((((((.((((((....(((((...............))))))))))).))))))).))))((((......-------------------------)))). ( -20.76) >DroYak_CAF1 2458 102 - 1 AGG-GACAAUUAAAUGAUGUUGCUAUAGAGUGUUUAAAUCACAACUUAUUCAAUAGUCUAUAGCAACACACUUUAA-GCCCAUAUG------------GGCUAUCAAUGUUGGCCA .((-(....(((((((.((((((((((((.((((.................)))).)))))))))))))).)))))-.))).....------------(((((.......))))). ( -32.93) >consensus AGG_GACAAUUAAAUUAUGUUGCUAUAGAGUAUCCAAAGCACAACUUAUUCAAUGGACUAUAGAAACAUAACUUAA_GCCCAUAUA____________GG__AUCAAUGUUGGCCA .........((((.((((((((((((((.............................)))))))))))))).)))).(((.((((.....................)))).))).. ( -9.16 = -12.38 + 3.23)


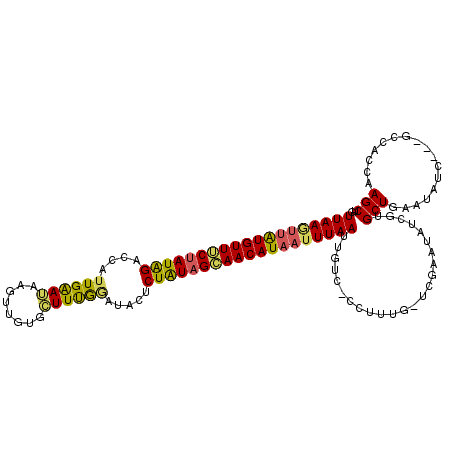
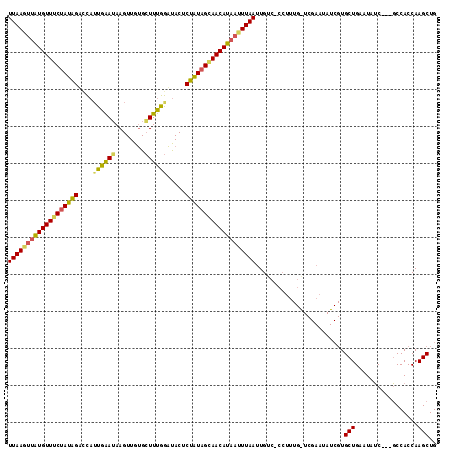
| Location | 19,458,752 – 19,458,855 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.09 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -18.21 |
| Energy contribution | -18.56 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19458752 103 + 22224390 UUAAGUUAUGUUUCUAUGGGUCAUUGGAGAAGUUGGGCUUCGGUUACGCUGUUGAAACAUAAGUUAAUUGGUCCCAU-------------UGCUGAAUAGCUCCUCCACCAAGCUG ((((.(((((((((.((((((.(((((((........))))))).)).)))).))))))))).))))(((((.....-------------.(((....)))......))))).... ( -27.90) >DroEre_CAF1 2577 112 + 1 UUAAGUUAUGUUUCUAUAGUCCAAUGAAUAAGUUGUGCUUUGGAUACUCUAUAGCAACAUAAUUUAAUUGUC-CAUUUGAUCGAAUAUCGUGCUGAAUAUC---GCCACCAAGCUG ((((((((((((.(((((((((((...((.....))...)))))....)))))).)))))))))))).....-................(((.((.....)---).)))....... ( -22.00) >DroYak_CAF1 2485 112 + 1 UUAAAGUGUGUUGCUAUAGACUAUUGAAUAAGUUGUGAUUUAAACACUCUAUAGCAACAUCAUUUAAUUGUC-CCUUUGGUCGAAUAUCGUGCUGAAUAUC---GCCACCAAGCUG ...(((((((((((((((((...((((((........))))))....)))))))))))).))))).......-..(((((((((...((.....))...))---)..))))))... ( -26.20) >consensus UUAAGUUAUGUUUCUAUAGACCAUUGAAUAAGUUGUGCUUUGGAUACUCUAUAGCAACAUAAUUUAAUUGUC_CCUUUG_UCGAAUAUCGUGCUGAAUAUC___GCCACCAAGCUG (((((((((((((((((((....((((((........)))))).....)))))))))))))))))))........................(((.................))).. (-18.21 = -18.56 + 0.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:59 2006