
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,441,184 – 19,441,287 |
| Length | 103 |
| Max. P | 0.968075 |

| Location | 19,441,184 – 19,441,287 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.47 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -16.31 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.50 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
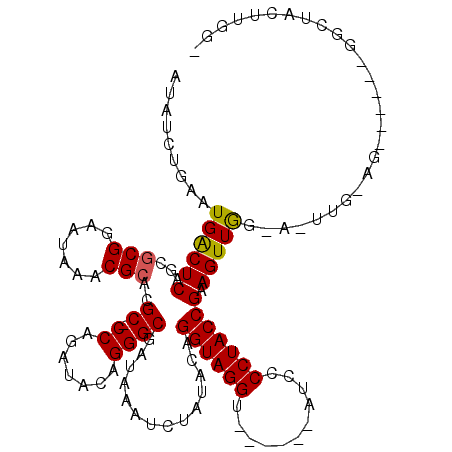
>X_DroMel_CAF1 19441184 103 + 22224390 AUAUCUGAAUGACUCAGCACGGAAUAAACGCACGCCCCAUAUACAGGGCAAUAAAUCCAUACAGGUAGGUGUAUCUAUGCCCUACCGAAGUUGCCACUUGAAG----------------- ....((((.....))))...((....(((....((((........))))..............((((((.(((....)))))))))...))).))........----------------- ( -25.10) >DroEre_CAF1 12780 98 + 1 AUAUCUGAAUGACUCAGCGCGGAAUAAACGCGCGC-CCAGAUACAGGGCGAUAUAUCUAUACAGGUAGGU------UUCCCCUACCGAAGUUAG---------------GGCUACUUGGA (((((...........(((((.......)))))((-((.......))))))))).......(((((((..------...(((((.......)))---------------))))))))).. ( -33.10) >DroYak_CAF1 13133 113 + 1 AUAUCCGAAUGGCUCAGCGCGGAAUAAACGCAGGC-CCAGAUACAGGGCGGUAAAUCUAUACAGGUAGGU------CUCCCCUACCGAAGUUGGGAUUUGGAGCGCUUUGGCUACUUGGU ....((((.(((((.(((((.............((-((.......))))..(((((((..((.((((((.------....))))))...))..)))))))..)))))..))))).)))). ( -40.20) >consensus AUAUCUGAAUGACUCAGCGCGGAAUAAACGCACGC_CCAGAUACAGGGCGAUAAAUCUAUACAGGUAGGU______AUCCCCUACCGAAGUUGG_A_UUG_AG______GGCUACUUGG_ .........((((((...(((.......)))..((.((.......))))..............((((((...........))))))).)))))........................... (-16.31 = -16.20 + -0.11)

| Location | 19,441,184 – 19,441,287 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.47 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -19.61 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
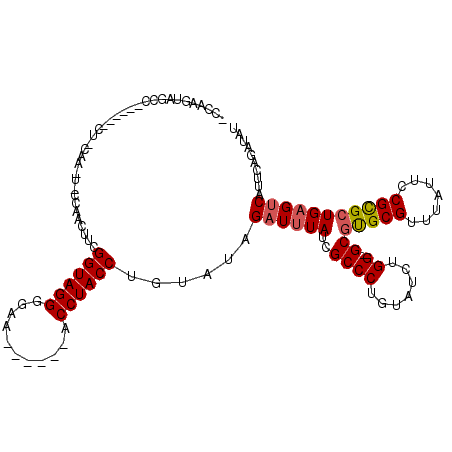
>X_DroMel_CAF1 19441184 103 - 22224390 -----------------CUUCAAGUGGCAACUUCGGUAGGGCAUAGAUACACCUACCUGUAUGGAUUUAUUGCCCUGUAUAUGGGGCGUGCGUUUAUUCCGUGCUGAGUCAUUCAGAUAU -----------------.....((((((....((((((.((.(((((..(((...((.....)).......(((((......))))))))..))))).)).))))))))))))....... ( -32.50) >DroEre_CAF1 12780 98 - 1 UCCAAGUAGCC---------------CUAACUUCGGUAGGGGAA------ACCUACCUGUAUAGAUAUAUCGCCCUGUAUCUGG-GCGCGCGUUUAUUCCGCGCUGAGUCAUUCAGAUAU ........(((---------------(.......((((((....------.)))))).....(((((((......)))))))))-))(((((.......)))))...(((.....))).. ( -32.00) >DroYak_CAF1 13133 113 - 1 ACCAAGUAGCCAAAGCGCUCCAAAUCCCAACUUCGGUAGGGGAG------ACCUACCUGUAUAGAUUUACCGCCCUGUAUCUGG-GCCUGCGUUUAUUCCGCGCUGAGCCAUUCGGAUAU .((.(((.((...(((((...(((((...((...((((((....------.)))))).))...)))))...((((.......))-)).............)))))..)).))).)).... ( -33.90) >consensus _CCAAGUAGCC______CU_CAA_U_CCAACUUCGGUAGGGGAA______ACCUACCUGUAUAGAUUUAUCGCCCUGUAUCUGG_GCGUGCGUUUAUUCCGCGCUGAGUCAUUCAGAUAU ..................................((((((...........))))))......((((((..((((.......)).))(((((.......))))))))))).......... (-19.61 = -20.17 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:54 2006