
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,436,411 – 19,436,546 |
| Length | 135 |
| Max. P | 0.988619 |

| Location | 19,436,411 – 19,436,517 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -16.17 |
| Energy contribution | -15.73 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
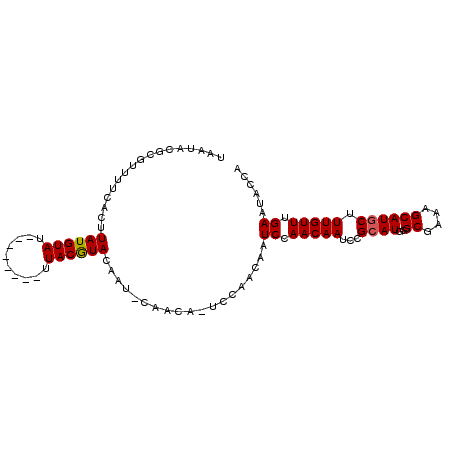
>X_DroMel_CAF1 19436411 106 + 22224390 UAAAUAGUGCCCGCACAGGCGCUGCACUCGUACGUAUACGUAAUACGCGUUUACACUUAUGUACAUAAAUAUGUACAUACAU-------GUCCAACAAUCCAACAAUCCGCAU .....((((((......))))))((.......(((((.....))))).(((.(((..(((((((((....)))))))))..)-------))..))).............)).. ( -25.80) >DroSec_CAF1 4156 97 + 1 UAAAUAGUGCCCGCACAAGCGCUGCACUCGCACGUAUACGUAAUACGCGUUUUCACGUAUGUAU--------UUACGUACAAUUCAACA--------AUCCAACAAUCCGCAU .....(((((.(((....)))..))))).((.(((((.....))))).........((((((..--------..)))))).........--------............)).. ( -20.10) >DroSim_CAF1 4513 105 + 1 UAAAUAGUGCCCGCACAAGCGCUGCACUCGUACGUAUACGUAAUACGCGUUUUCACUUAUGUAU--------UUACGUACAAUCCAACAAUCCAACAAUCCAACAAUCCGCAU .....(((((.(((....)))..))))).((((((((((((((.............))))))).--------.)))))))................................. ( -20.92) >consensus UAAAUAGUGCCCGCACAAGCGCUGCACUCGUACGUAUACGUAAUACGCGUUUUCACUUAUGUAU________UUACGUACAAU_CAACA_UCCAACAAUCCAACAAUCCGCAU .....(((((.(((....)))..))))).((.(((((.....)))))..........((((((..........))))))..............................)).. (-16.17 = -15.73 + -0.44)


| Location | 19,436,411 – 19,436,517 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -23.31 |
| Energy contribution | -22.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19436411 106 - 22224390 AUGCGGAUUGUUGGAUUGUUGGAC-------AUGUAUGUACAUAUUUAUGUACAUAAGUGUAAACGCGUAUUACGUAUACGUACGAGUGCAGCGCCUGUGCGGGCACUAUUUA .........((((.((((((..((-------((.(((((((((....))))))))).)))).)))((((((.....))))))...))).))))((((....))))........ ( -32.70) >DroSec_CAF1 4156 97 - 1 AUGCGGAUUGUUGGAU--------UGUUGAAUUGUACGUAA--------AUACAUACGUGAAAACGCGUAUUACGUAUACGUGCGAGUGCAGCGCUUGUGCGGGCACUAUUUA .(((............--------........((((((((.--------....(((((((....)))))))))))))))((..((((((...))))))..)).)))....... ( -28.40) >DroSim_CAF1 4513 105 - 1 AUGCGGAUUGUUGGAUUGUUGGAUUGUUGGAUUGUACGUAA--------AUACAUAAGUGAAAACGCGUAUUACGUAUACGUACGAGUGCAGCGCUUGUGCGGGCACUAUUUA .(((............................(((((((((--------.(((....((....))..))))))))))))((((((((((...)))))))))).)))....... ( -27.10) >consensus AUGCGGAUUGUUGGAUUGUUGGA_UGUUG_AUUGUACGUAA________AUACAUAAGUGAAAACGCGUAUUACGUAUACGUACGAGUGCAGCGCUUGUGCGGGCACUAUUUA ................................((((((((.................((....))(((.....))).))))))))(((((..(((....))).)))))..... (-23.31 = -22.87 + -0.44)



| Location | 19,436,451 – 19,436,546 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.19 |
| Mean single sequence MFE | -17.83 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19436451 95 + 22224390 UAAUACGCGUUUACACUUAUGUACAUAAAUAUGUACAUACAU-------GUCCAACAAUCCAACAAUCCGCAUGGGCGAAAGCAUACUUUGUUUGAAUACCA ........(((.(((..(((((((((....)))))))))..)-------))..)))..((.(((((.((....))((....)).....))))).))...... ( -20.40) >DroSec_CAF1 4196 86 + 1 UAAUACGCGUUUUCACGUAUGUAU--------UUACGUACAAUUCAACA--------AUCCAACAAUCCGCAUGGGCGAAAGCAUGCUUUGUUUGAAUACCA ......(.((.((((.((((((..--------..)))))).........--------....(((((...((((..((....)))))).))))))))).))). ( -18.20) >DroSim_CAF1 4553 94 + 1 UAAUACGCGUUUUCACUUAUGUAU--------UUACGUACAAUCCAACAAUCCAACAAUCCAACAAUCCGCAUGGGCGAAAGCAUGCUUUGUUUGAAUACCA ......(.((.((((....(((((--------....)))))....................(((((...((((..((....)))))).))))))))).))). ( -14.90) >consensus UAAUACGCGUUUUCACUUAUGUAU________UUACGUACAAU_CAACA_UCCAACAAUCCAACAAUCCGCAUGGGCGAAAGCAUGCUUUGUUUGAAUACCA .................((((((..........))))))...................((.(((((...((((..((....)))))).))))).))...... (-12.32 = -12.43 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:50 2006