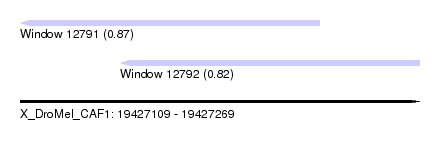
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,427,109 – 19,427,269 |
| Length | 160 |
| Max. P | 0.865190 |

| Location | 19,427,109 – 19,427,229 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -42.28 |
| Energy contribution | -41.96 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
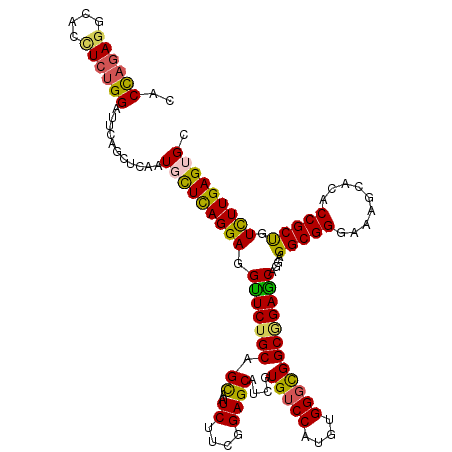
>X_DroMel_CAF1 19427109 120 - 22224390 UUCUGCAGCAAUCUUCGGAGCAUUGUGUCCAUGUGGUCGGCUGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGCCAAAAAGGUGGGAAUACUCGAUGCCAACUGAAGCUUGCUG .....((((((.((((((.((((((.((((((.(..(.(((....((((((((((.........))))).)))))...))).)..).))))....)).))))))...)))))).)))))) ( -46.20) >DroSec_CAF1 25420 120 - 1 UUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGCGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGCCAGAUAGGUGGAGAUACUAGGUGCCAGCUGAAGCUUGCUG .....((((((.((((((.(((((((.(((((.((...((((...((((((((((.........))))).)))))..))))...)).))))).))....)))))...)))))).)))))) ( -49.50) >DroSim_CAF1 21507 120 - 1 CUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGUGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGGGCCAGAUAGGUGGAGAUACUAGGUGCCAGCUGAAGCUUGCUG .....((((((.((((((.(((((((.(((((.((..((((....((((((((((.........))))).)))))...))))..)).))))).))....)))))...)))))).)))))) ( -51.30) >DroEre_CAF1 20176 120 - 1 UUCUGCAGUAAUCUUCGGAGCAUCAUGUCCAUGUGGGCGGCGGAACAGGAGGCGGGAAAGCAGACCGCUGUUUUGAGUGCCAGAAAGGUGGAGAUACUGGGUGCCAGUUGGAGCUUGCUG .....((((((.((((((.(((((((.(((((.(.((((.(((((((....((((.........)))))))))))..))))....).))))).))....)))))...)))))).)))))) ( -41.70) >DroYak_CAF1 26325 120 - 1 UUCUGCAGUAAUCUCCGGAGCAUCGUGUCCAUGUGGGCGGCAGAGCAGGAGGCGGGAAAGCAGACCGCCGUCUUGAGGGACAGAUAGGCGGAGAAACUUGGUGCCACUUGAAGCUUGCUG .....((((((((((((....(((.(((((....(((((((...((.....))((.........)))))))))....))))))))...))))))........((........)))))))) ( -39.80) >consensus UUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGCGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGCCAGAUAGGUGGAGAUACUAGGUGCCAGCUGAAGCUUGCUG .....((((((.((((((.(((((((.(((((.((...(((....((((((((((.........))))).)))))...)))...)).))))).))....)))))...)))))).)))))) (-42.28 = -41.96 + -0.32)



| Location | 19,427,149 – 19,427,269 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -47.00 |
| Consensus MFE | -40.22 |
| Energy contribution | -39.78 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19427149 120 - 22224390 CACUAGAGGCACAUCUGGAUUCAGCUCAAUGCUUAGGAGGUUCUGCAGCAAUCUUCGGAGCAUUGUGUCCAUGUGGUCGGCUGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGC ((((.(((((...((((..(((((((((((((((.((((((((....).))))))).)))))))).(.((....)).)))))))))))).(((((.........)))))))))).)))). ( -46.20) >DroSec_CAF1 25460 120 - 1 CACCAGAGUCACUUCUGGAUUCAGCUCAAUGCUCAGGAGGUUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGCGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGC ..((((((....))))))(((((((((.((((((.((((((((....).))))))).))))))(((((((....)))).)))))))((..(((((.........)))))..))))))).. ( -47.20) >DroSim_CAF1 21547 120 - 1 CACCAGAGUCACCUCUGGAUUCAGCUCAAUGCUCAGGAGGCUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGUGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGGGC ..((((((....)))))).............(((((((.(((((((.((..((....))))....(..((....))..))))))))....(((((.........))))).)))))))... ( -45.20) >DroEre_CAF1 20216 120 - 1 CACCAGAGGCACCUCUGGAUUCAGCUCAAUGCUUAGGAGGUUCUGCAGUAAUCUUCGGAGCAUCAUGUCCAUGUGGGCGGCGGAACAGGAGGCGGGAAAGCAGACCGCUGUUUUGAGUGC ..((((((....)))))).....((((((.((.......(((((((.....((....))......(((((....))))))))))))....(((((.........))))))).)))))).. ( -45.40) >DroYak_CAF1 26365 120 - 1 CACCACAGGCACCUCAGGAUUCAGCUCAAUGUUCAGGAGGUUCUGCAGUAAUCUCCGGAGCAUCGUGUCCAUGUGGGCGGCAGAGCAGGAGGCGGGAAAGCAGACCGCCGUCUUGAGGGA ...........(((((((((...((((.((((((.((((((((....).))))))).)))))).((((((....)))).)).))))....(((((.........)))))))))))))).. ( -51.00) >consensus CACCAGAGGCACCUCUGGAUUCAGCUCAAUGCUCAGGAGGUUCUGCAGCAAUCUUCGGAGCAUCGUGUCCAUGUGGGCGGCGGAGCAGGAGGCGGGAAAGCACACCGCUGUCUUGAGUGC ..((((((....))))))...........(((((((((.(((((((.((..((....))))....(((((....))))))))))))....(((((.........))))).))))))))). (-40.22 = -39.78 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:46 2006