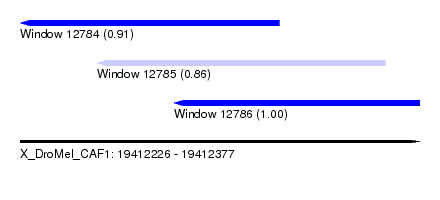
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,412,226 – 19,412,377 |
| Length | 151 |
| Max. P | 0.995651 |

| Location | 19,412,226 – 19,412,324 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -29.34 |
| Energy contribution | -29.23 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19412226 98 - 22224390 AGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCAUACUCCAAUGUAUGCAU--GUG--UGUGUGUGUAUUUGGAUUCGCAACGGCGAUAAAUGCACCC ........(((.....((...(((((((((.((((((((((......))))))))--)).--)))))))))....))..((((....))))....))).... ( -38.90) >DroSec_CAF1 12065 100 - 1 AGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACACUCCGAUGUAUGCGUCUGUG--UGUGUGUGUAUGUGGAUUCGCAACGGCGAUAAAUGCACCC ........(((.......((((((((((((.(((((((......((((....))))))))--))).)))))))))))).((((....))))....))).... ( -38.00) >DroYak_CAF1 12148 102 - 1 AGACGCCCGCAAAACGCCUCCGCAUGCACACACAUGCACACUCCGAUAUAUAUGUGUGUGUGUGUAUGAGUGUGUGGAUUCGCAACGGCGACAAAUGCACCC ........(((.......(((((((((.(((((((((((((............))))))))))))..).))))))))).((((....))))....))).... ( -35.60) >consensus AGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACACUCCGAUGUAUGCGU_UGUG__UGUGUGUGUAUGUGGAUUCGCAACGGCGAUAAAUGCACCC ........(((.......((((((((((((.(((..((((.....(((....))).))))..))).)))))))))))).((((....))))....))).... (-29.34 = -29.23 + -0.10)



| Location | 19,412,255 – 19,412,364 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -48.49 |
| Consensus MFE | -35.04 |
| Energy contribution | -34.94 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19412255 109 - 22224390 CGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCAUACUCCAAUGUAUGCAU--GUG--UGUGUGUGUAU .((((((........((((((.......((((....)))))))))).........))))))(((((((((.((((((((((......))))))))--)).--))))))))).. ( -54.74) >DroSec_CAF1 12094 111 - 1 CGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGAAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACACUCCGAUGUAUGCGUCUGUG--UGUGUGUGUAU .((((((........((((((.......(((......))))))))).........))))))((((((((((((..(((.((......)).))))))))))--)))))...... ( -42.94) >DroYak_CAF1 12177 113 - 1 CGGGGGCCAAAAUUCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACACACAUGCACACUCCGAUAUAUAUGUGUGUGUGUGUAUGAGUGU (((((((........((((((.......((((....)))))))))).........)))))))((((((((((((((((..............))))))))))))))))..... ( -47.78) >consensus CGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACACUCCGAUGUAUGCGU_UGUG__UGUGUGUGUAU (((((((........((((((.......((((....)))))))))).........))))))).(((((((.(((..((((.....(((....))).))))..))).))))))) (-35.04 = -34.94 + -0.10)


| Location | 19,412,284 – 19,412,377 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -37.54 |
| Consensus MFE | -35.87 |
| Energy contribution | -36.74 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19412284 93 - 22224390 GCCGGAGCUCCGACGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCAUA ..(((....)))..((((((........((((((.......((((....)))))))))).........))))))(((((......)))))... ( -35.04) >DroSec_CAF1 12125 93 - 1 GCCGGAGCUCCGGCGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGAAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACA (((((....)))))((((((........((((((.......(((......))))))))).........))))))(((((......)))))... ( -38.34) >DroEre_CAF1 12019 84 - 1 GCCGGAGCUCCAGCGGAGGCCAAAAUCCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAU---------UGCACA ((.((....)).((((((((........((((((.......((((....)))))))))).........))))))))..---------.))... ( -34.74) >DroYak_CAF1 12210 93 - 1 GCCGGAGCUCCGGCGGGGGCCAAAAUUCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACACACAUGCACA (((((....)))))((((((........((((((.......((((....)))))))))).........))))))(((((......)))))... ( -42.04) >consensus GCCGGAGCUCCGGCGGGGGCCAAAAUCCGCGUCUUUAGCCGGCGGAUAUCCGCAGACGCCCGCAAAACGCCUCCGCAUGCACAGACAUGCACA (((((....)))))((((((........((((((.......((((....)))))))))).........))))))(((((......)))))... (-35.87 = -36.74 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:40 2006