| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,376,094 – 19,376,211 |
| Length | 117 |
| Max. P | 0.860257 |

| Location | 19,376,094 – 19,376,211 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -22.74 |
| Energy contribution | -21.94 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19376094 117 + 22224390 AUAAGAUUGGAUAAGAUUGUGUAAGAUAGGGUUGUUAUAUAAAUUUCUUCAAACCGAAAGUCUUUGUUAGUUUGAUUAGAAUUCUAUGCGAUGAAAUUUGUAUAGCAUCCUU---UGAGU ...........................((((.(((((((((((((((.((((((((((....))))...)))))).(((....)))......))))))))))))))).))))---..... ( -24.10) >DroSec_CAF1 9175 112 + 1 AUGAGAUUGGAAAAGAUUGUGU-----AGAGGUGUUACAUAAAUUUCUUCAAGCCGAAGGUCUUUGUUAAUUUGAUUAGAAUUCUGUGCGAAGAAAUUUUUAUAGCAUCUUA---AGAGC (..(..((....))..)..)..-----.(((((((((...((((((((((..((.(((..(((..(((.....))).))).))).))..))))))))))...))))))))).---..... ( -25.30) >DroSim_CAF1 5548 112 + 1 AUGAGAUUGGAAAAGAUUGUGU-----AGAGGUGUUACAUAAAUUUCUUCAAACCGAAGGUCUUUGUUAAUUUGAUUAGAAUUCUGUGCGAAGAAAUUUUUAUAGCAUCUUA---AGAGC (..(..((....))..)..)..-----.(((((((((...((((((((((..((.(((..(((..(((.....))).))).))).))..))))))))))...))))))))).---..... ( -25.00) >DroEre_CAF1 8782 115 + 1 AUGAGGUUGGAUGAGAUUUUAC-----AGGGCUGUUACAUAAAUCUCUUCAAGCCGAAGGUCUAUGUAAGUUUGGUUGGAAUUCUGUGAGAAGAGAUUUCUAUAGCAUCCUGAAUACAGU .(((((((......)))))))(-----((((.(((((.(.((((((((((..((.(((..((((............)))).))).))..)))))))))).).))))))))))........ ( -30.70) >DroYak_CAF1 8874 115 + 1 AUGAGGUUGGAUGAGAUUGUAC-----AGGGGUGUUACAUAAAUUUCUUCAAGCCGAAAGUCUAUGUAAAUUUGGUUGGAAUUCGGUGAGAAGAGAUUUCUAUAGCACUCUGAAUAGAGC ................((((..-----.(((((((((.(.((((((((((..((((((..((((............)))).))))))..)))))))))).).)))))))))..))))... ( -31.90) >consensus AUGAGAUUGGAUAAGAUUGUGU_____AGGGGUGUUACAUAAAUUUCUUCAAGCCGAAGGUCUUUGUUAAUUUGAUUAGAAUUCUGUGCGAAGAAAUUUCUAUAGCAUCCUA___AGAGC ............................(((((((((...((((((((((..((.(((..(((..............))).))).))..))))))))))...)))))))))......... (-22.74 = -21.94 + -0.80)

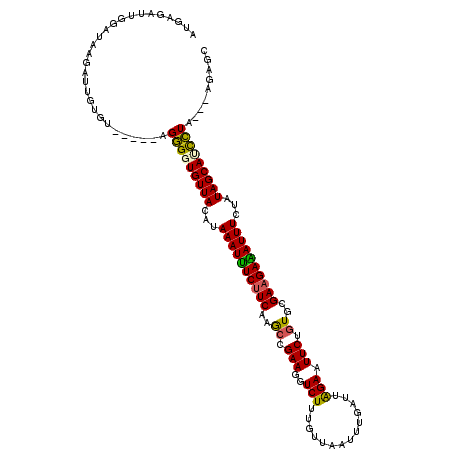

| Location | 19,376,094 – 19,376,211 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -17.12 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.01 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19376094 117 - 22224390 ACUCA---AAGGAUGCUAUACAAAUUUCAUCGCAUAGAAUUCUAAUCAAACUAACAAAGACUUUCGGUUUGAAGAAAUUUAUAUAACAACCCUAUCUUACACAAUCUUAUCCAAUCUUAU .....---..((.((.((((.(((((((......(((....))).(((((((.............))))))).))))))).)))).)).))............................. ( -15.82) >DroSec_CAF1 9175 112 - 1 GCUCU---UAAGAUGCUAUAAAAAUUUCUUCGCACAGAAUUCUAAUCAAAUUAACAAAGACCUUCGGCUUGAAGAAAUUUAUGUAACACCUCU-----ACACAAUCUUUUCCAAUCUCAU .....---..(((((.((((.((((((((((((...(((.(((..............)))..))).))..)))))))))).)))).))..)))-----...................... ( -14.54) >DroSim_CAF1 5548 112 - 1 GCUCU---UAAGAUGCUAUAAAAAUUUCUUCGCACAGAAUUCUAAUCAAAUUAACAAAGACCUUCGGUUUGAAGAAAUUUAUGUAACACCUCU-----ACACAAUCUUUUCCAAUCUCAU .....---..(((((.((((.(((((((((((.((.(((.(((..............)))..))).)).))))))))))).)))).))..)))-----...................... ( -15.94) >DroEre_CAF1 8782 115 - 1 ACUGUAUUCAGGAUGCUAUAGAAAUCUCUUCUCACAGAAUUCCAACCAAACUUACAUAGACCUUCGGCUUGAAGAGAUUUAUGUAACAGCCCU-----GUAAAAUCUCAUCCAACCUCAU ........((((.((.((((.((((((((((.....(((.......................))).....)))))))))).)))).))..)))-----)..................... ( -20.50) >DroYak_CAF1 8874 115 - 1 GCUCUAUUCAGAGUGCUAUAGAAAUCUCUUCUCACCGAAUUCCAACCAAAUUUACAUAGACUUUCGGCUUGAAGAAAUUUAUGUAACACCCCU-----GUACAAUCUCAUCCAACCUCAU ........(((.(((.((((.((((.(((((...(((((.......................)))))...))))).)))).)))).)))..))-----)..................... ( -18.80) >consensus GCUCU___CAGGAUGCUAUAAAAAUUUCUUCGCACAGAAUUCUAAUCAAAUUAACAAAGACCUUCGGCUUGAAGAAAUUUAUGUAACACCCCU_____ACACAAUCUUAUCCAAUCUCAU ..........((.((.((((.((((((((((.....(((.((................))..))).....)))))))))).)))).)).))............................. (-12.29 = -12.01 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:28 2006