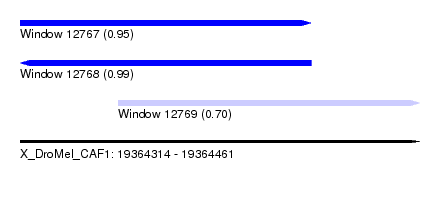
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,364,314 – 19,364,461 |
| Length | 147 |
| Max. P | 0.991903 |

| Location | 19,364,314 – 19,364,421 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -17.15 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19364314 107 + 22224390 CAUCAUUUUUUUUUUUUUUUCGUGACAUUACCUAGCAUAUAACAAGUAAGUGGGGUAGCCCUGCUGCCAACGCAGCUACCCCAGCUAAUACAAUCUGGAAUAUCACC .....................((((.(((.((((((.(((.....)))..(((((((((..(((.......)))))))))))))))).........))))).)))). ( -26.20) >DroSec_CAF1 7245 102 + 1 CAUCAUUUUUU-----UUUUCUUGACAUUACCUAGCAUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCACAGCUACCCUGCCUAAUACAAUCUGGAAUAUCACC ...........-----..((((.((......((((..(((.....))).))))(((((....((((......))))....)))))........)).))))....... ( -16.00) >DroSim_CAF1 7124 104 + 1 CAUCAUUUUUU---UUUUUUCGUGACAUUACCUAGCAUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCGCAGCUACCCCGCCUAAUACAAUCUGGAAUAUCACC ...........---.......((((.(((.((.....(((.....)))...((((((((..(((.......)))))))))))..............))))).)))). ( -23.10) >DroEre_CAF1 7270 88 + 1 ---------UU---UUUUUUCGUGACAUUACUUAGCAUAUAACA----AUUGGGGUAGCCCU---GCCACCGCAGCUACCCCACGUAAUACAAUCUGGAAUAUCACC ---------..---.......((((.(((.((.....(((.((.----..(((((((((..(---((....)))))))))))).)).)))......))))).)))). ( -23.00) >consensus CAUCAUUUUUU___UUUUUUCGUGACAUUACCUAGCAUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCGCAGCUACCCCACCUAAUACAAUCUGGAAUAUCACC .....................((((.(((.((.((...............(((((((((.....(((....))))))))))))...........))))))).)))). (-17.15 = -17.27 + 0.12)



| Location | 19,364,314 – 19,364,421 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.88 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19364314 107 - 22224390 GGUGAUAUUCCAGAUUGUAUUAGCUGGGGUAGCUGCGUUGGCAGCAGGGCUACCCCACUUACUUGUUAUAUGCUAGGUAAUGUCACGAAAAAAAAAAAAAAAUGAUG .((((((((((((..((((..((.((((((((((((.......))..))))))))))....))...))))..)).)).))))))))..................... ( -32.60) >DroSec_CAF1 7245 102 - 1 GGUGAUAUUCCAGAUUGUAUUAGGCAGGGUAGCUGUGAUGGCAGCAGGGCUACCCCAGUUACUUGUUAUAUGCUAGGUAAUGUCAAGAAAA-----AAAAAAUGAUG ..(((((((((((..((((.((((..((((((((.((.......)).))))))))......)))).))))..)).)).)))))))......-----........... ( -27.20) >DroSim_CAF1 7124 104 - 1 GGUGAUAUUCCAGAUUGUAUUAGGCGGGGUAGCUGCGAUGGCAGCAGGGCUACCCCAGUUACUUGUUAUAUGCUAGGUAAUGUCACGAAAAAA---AAAAAAUGAUG .((((((((((((..((((.((((.(((((((((((.......))..))))))))).....)))).))))..)).)).)))))))).......---........... ( -32.70) >DroEre_CAF1 7270 88 - 1 GGUGAUAUUCCAGAUUGUAUUACGUGGGGUAGCUGCGGUGGC---AGGGCUACCCCAAU----UGUUAUAUGCUAAGUAAUGUCACGAAAAAA---AA--------- .((((((((.(.....((((.((.((((((((((((....))---..))))))))))..----.))...))))...).)))))))).......---..--------- ( -30.10) >consensus GGUGAUAUUCCAGAUUGUAUUAGGCGGGGUAGCUGCGAUGGCAGCAGGGCUACCCCAGUUACUUGUUAUAUGCUAGGUAAUGUCACGAAAAAA___AAAAAAUGAUG .((((((((((((..((((.((((((((((((((((....)).....))))))))))....)))).))))..)).)).))))))))..................... (-25.06 = -26.88 + 1.81)



| Location | 19,364,350 – 19,364,461 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19364350 111 + 22224390 AUAUAACAAGUAAGUGGGGUAGCCCUGCUGCCAACGCAGCUACCCCAGCUAAUACAAUCUGGAAUAUCACCCAUCCGGCCUCACCACUUCGGCACUUCGGCAGGGAGUUUC ....(((..((((((((((((((..(((.......)))))))))))).))..)))..............(((..((((((..........)))....)))..))).))).. ( -31.10) >DroSec_CAF1 7276 111 + 1 AUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCACAGCUACCCUGCCUAAUACAAUCUGGAAUAUCACCCAUCCGGCCUCACCACUUCGGCACUUCGGCGAGUAGUUUC ..............(((((((((...)))))).))).((((((..((((..........(((........)))....(((..........))).....)))).)))))).. ( -24.50) >DroSim_CAF1 7157 111 + 1 AUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCGCAGCUACCCCGCCUAAUACAAUCUGGAAUAUCACCCAUCCGGCCUCACCACUUCGGCACUUCGGCGAGUAGUUUC ............(((((((((((..(((.......)))))))))))(((..........(((........)))....(((..........))).....))).)))...... ( -29.50) >DroEre_CAF1 7294 101 + 1 AUAUAACA----AUUGGGGUAGCCCU---GCCACCGCAGCUACCCCACGUAAUACAAUCUGGAAUAUCACCCAUCCGGCCUCACCACA---CCACUCCAUCAACGAGUUUC ........----..(((((((((..(---((....))))))))))))...........(((((..........)))))..........---..((((.......))))... ( -23.80) >consensus AUAUAACAAGUAACUGGGGUAGCCCUGCUGCCAUCGCAGCUACCCCACCUAAUACAAUCUGGAAUAUCACCCAUCCGGCCUCACCACUUCGGCACUUCGGCAAGGAGUUUC ..............(((((((((.....(((....))))))))))))...........(((((..........)))))...............(((((.....)))))... (-19.00 = -19.75 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:25 2006