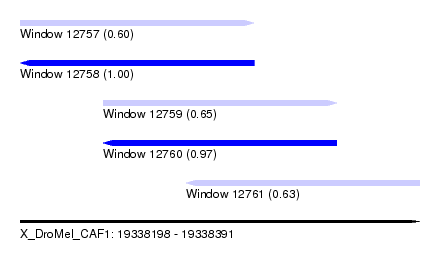
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,338,198 – 19,338,391 |
| Length | 193 |
| Max. P | 0.995518 |

| Location | 19,338,198 – 19,338,311 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -15.88 |
| Energy contribution | -16.88 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19338198 113 + 22224390 GUUACAUUUCGUGCCUAUAGCUCUAUUAGCUACAUGCUAUAUGCUAUAUGCUAUAUGCCAUAUGCCAUAUGCCAUUAGCU-------AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUU ..........((((..((.((.....((((((.(((.((((((.(((((((.....).)))))).)))))).))))))))-------)...)).))..)))).((((((...)))))).. ( -32.30) >DroSec_CAF1 10896 106 + 1 GUUACGUUUCGUGCCUAUAGCUCUAUUAGCUACAUGCUAUAUG-------CUAUAUGCCAUGUGCCAUAUGACAUUAGCU-------AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUU ..........((((..((.((.....((((((.(((.((((((-------.(((((...))))).)))))).))))))))-------)...)).))..)))).((((((...)))))).. ( -28.00) >DroEre_CAF1 10617 100 + 1 GUUACAUUUCGUGCCUAUAGCUCUAUUAGCUACAUA-----UG--------UAUAUGCUAUAUGCUAUAUGCCAUUAGCU-------UUUUGCCAUUAGCGCCGGCGUCGACGACGCUUU ..........((((..((.((...(..(((((....-----.(--------(((((((.....)).))))))...)))))-------..).)).))..)))).((((((...)))))).. ( -24.60) >DroYak_CAF1 10667 99 + 1 GUUACAUUCCGUGCCUUUAGCUCUAUUAGCU---------------------AUAUGCUAUAUGCCAUAUGCUAUUAGCUAUUUGCCAUUUGCCAUUAGCGCCGGCGUCGACGACGCUUU ..........((((...(((((....((((.---------------------((((((.....).)))))))))..)))))...((.....)).....)))).((((((...)))))).. ( -23.10) >consensus GUUACAUUUCGUGCCUAUAGCUCUAUUAGCUACAUG_____UG________UAUAUGCCAUAUGCCAUAUGCCAUUAGCU_______AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUU ..........((((...(((((.....)))))........................((((...((.....))...))))...................)))).((((((...)))))).. (-15.88 = -16.88 + 1.00)


| Location | 19,338,198 – 19,338,311 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.45 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19338198 113 - 22224390 AAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU-------AGCUAAUGGCAUAUGGCAUAUGGCAUAUAGCAUAUAGCAUAUAGCAUGUAGCUAAUAGAGCUAUAGGCACGAAAUGUAAC ...((((..(((..(((.((((((.((((....-------.)))).))))(((((.(....).))))).)).((((((.((((((.....))).)))..))))))))).))).))))... ( -34.60) >DroSec_CAF1 10896 106 - 1 AAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU-------AGCUAAUGUCAUAUGGCACAUGGCAUAUAG-------CAUAUAGCAUGUAGCUAAUAGAGCUAUAGGCACGAAACGUAAC ...((((..(((..(((...((((.((((....-------.))))(((..(((((.(....).)))))..-------))).)))).(((((((.....)))))))))).))).))))... ( -34.40) >DroEre_CAF1 10617 100 - 1 AAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAA-------AGCUAAUGGCAUAUAGCAUAUAGCAUAUA--------CA-----UAUGUAGCUAAUAGAGCUAUAGGCACGAAAUGUAAC ...(((((...)))))((((((((.((((....-------.)))).)))).((((((.((((((((((.--------..-----.)))).))).)))..)))))).)).))......... ( -28.80) >DroYak_CAF1 10667 99 - 1 AAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAUGGCAAAUAGCUAAUAGCAUAUGGCAUAUAGCAUAU---------------------AGCUAAUAGAGCUAAAGGCACGGAAUGUAAC ..(((((((.......)))))))....(((..((((...(((((..(((((((((........)))))---------------------.))))....)))))...)).))...)))... ( -27.30) >consensus AAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU_______AGCUAAUGGCAUAUGGCAUAUAGCAUAUA________CA_____CAUGUAGCUAAUAGAGCUAUAGGCACGAAAUGUAAC ...((((..(((..(((.((((((.((((............)))).)))).((((...))))))......................(((((((.....)))))))))).))).))))... (-22.64 = -22.45 + -0.19)


| Location | 19,338,238 – 19,338,351 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -27.11 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19338238 113 + 22224390 AUGCUAUAUGCUAUAUGCCAUAUGCCAUAUGCCAUUAGCU-------AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUUUCUCUGCCCGCCGACGAUCGGGGUUCUCGGAAAUCGGCAC ..((((.(((.((((((.(....).)))))).))))))).-------...((((...(((.(((((((((.((..((........)).)).))))).)))).)))....(....))))). ( -33.70) >DroSec_CAF1 10936 106 + 1 AUG-------CUAUAUGCCAUGUGCCAUAUGACAUUAGCU-------AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUUUCUCUGCCCGCCGACGAUCGGGGUUCUCGGAAAUCGGCAC (((-------.((((((.(....).)))))).)))..(((-------(........))))(((((((((...))))).(((((..(..(.(((.....))).)..)..))))).)))).. ( -31.40) >DroEre_CAF1 10653 104 + 1 -UG--------UAUAUGCUAUAUGCUAUAUGCCAUUAGCU-------UUUUGCCAUUAGCGCCGGCGUCGACGACGCUUUUCUCUGCCCGCCGACGAUCGGCGUUCUCGGAAAUCGGCAC -.(--------(((((((.....)).)))))).....(((-------..((.((...(((((((((((((.((..((........)).)).))))).))))))))...)).))..))).. ( -35.80) >DroYak_CAF1 10698 108 + 1 ------------AUAUGCUAUAUGCCAUAUGCUAUUAGCUAUUUGCCAUUUGCCAUUAGCGCCGGCGUCGACGACGCUUUUCUCUGCCCGCCGACGAUCGGCGUUCUCGGAAAUCGGCAC ------------....((((...((.....))...))))....((((((((.((...(((((((((((((.((..((........)).)).))))).))))))))...)))))).)))). ( -38.50) >consensus _UG________UAUAUGCCAUAUGCCAUAUGCCAUUAGCU_______AUUUGCCAUUAGCGCCGGCGUCGACGACGCUUUUCUCUGCCCGCCGACGAUCGGCGUUCUCGGAAAUCGGCAC ......................((((....((.....)).............((...(((((((((((((.((..((........)).)).))))).))))))))...)).....)))). (-27.11 = -27.68 + 0.56)


| Location | 19,338,238 – 19,338,351 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.87 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19338238 113 - 22224390 GUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU-------AGCUAAUGGCAUAUGGCAUAUGGCAUAUAGCAUAUAGCAU ((((((....(((......(((..(((((((((((........)).))))))))).))).((((.((((....-------.)))).))))...)))....))))))...((.....)).. ( -39.00) >DroSec_CAF1 10936 106 - 1 GUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU-------AGCUAAUGUCAUAUGGCACAUGGCAUAUAG-------CAU ((((((.....((.......))..(((((((((((........)).))))))))).))))))...((((....-------.))))(((..(((((.(....).)))))..-------))) ( -36.70) >DroEre_CAF1 10653 104 - 1 GUGCCGAUUUCCGAGAACGCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAA-------AGCUAAUGGCAUAUAGCAUAUAGCAUAUA--------CA- ..((((.(((((..(..(((((.....)))))..).).)))).(((((...)))))))))((((.((((....-------.)))).))))((((.((.....)))))).--------..- ( -37.30) >DroYak_CAF1 10698 108 - 1 GUGCCGAUUUCCGAGAACGCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAUGGCAAAUAGCUAAUAGCAUAUGGCAUAUAGCAUAU------------ .((((.(((((((..(.(((((..(((((((((((........)).))))))))).))))).)..))).))))))))....((((...((.....))...))))....------------ ( -40.40) >consensus GUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU_______AGCUAAUGGCAUAUGGCAUAUAGCAUAUA________CA_ ((((((............((((..(((((((((((........)).))))))))).))))((((.((((............)))).))))...))))))..................... (-34.22 = -34.85 + 0.63)

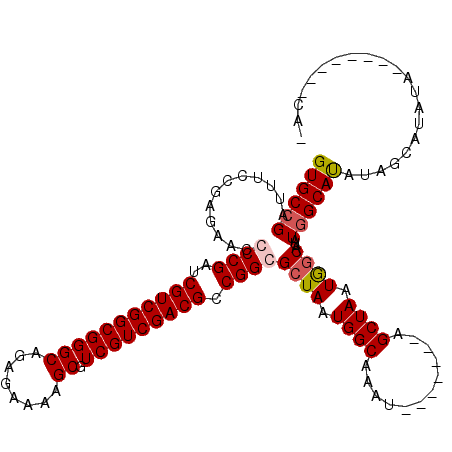
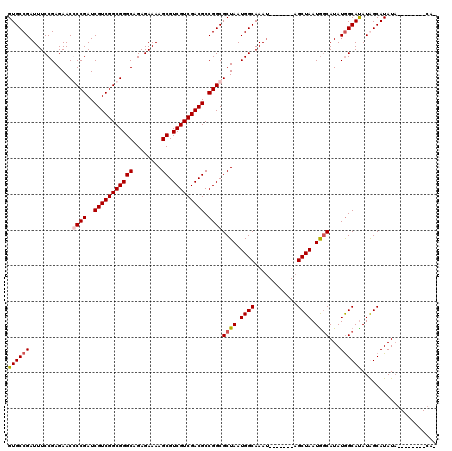
| Location | 19,338,278 – 19,338,391 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.86 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19338278 113 - 22224390 AAGCAGACACUGCGCUGAACGCUCAAACACUGAAAAACUAGUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU------- ..((.......((((((...((((....((((......))))((((((...((.......))...))))))))))........(((((...))))))))))).....))....------- ( -35.80) >DroSec_CAF1 10969 113 - 1 AAGCAGACACCGCGCUGAACGCUCAAACACUGAAAAACUAGUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU------- ..((.......((((((...((((....((((......))))((((((...((.......))...))))))))))........(((((...))))))))))).....))....------- ( -35.10) >DroEre_CAF1 10684 113 - 1 AAGCAGACACUGCGCUGAACGCUCAAACACUGAAAAACUAGUGCCGAUUUCCGAGAACGCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAA------- ..((((...))))(((......(((.....))).....((((((((.(((((..(..(((((.....)))))..).).)))).(((((...)))))))))))))..)))....------- ( -39.40) >DroYak_CAF1 10726 120 - 1 AAGCAGACACUGCGCUGAACGCUCAAACACUGAAAAACUAGUGCCGAUUUCCGAGAACGCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAUGGCAAAU ..((((...))))((((...(((((.....))).....((((((((.(((((..(..(((((.....)))))..).).)))).(((((...)))))))))))))...))...)))).... ( -40.60) >DroAna_CAF1 16766 96 - 1 AAGCUGACUCGGCGAAGAACGCUCCAAUACUGAAA-GCCAGCGCCGAAA--------------CUAGCGACUGGCUUUGAGACCCGGCCGCGACGCCGGCGACGACGGCGC--------- ..((((.(((((((.....))).......((.(((-((((((((.....--------------...))).)))))))).))..(((((......)))))))).).))))..--------- ( -38.40) >consensus AAGCAGACACUGCGCUGAACGCUCAAACACUGAAAAACUAGUGCCGAUUUCCGAGAACCCCGAUCGUCGGCGGGCAGAGAAAAGCGUCGUCGACGCCGGCGCUAAUGGCAAAU_______ ..((.......(((.....)))................((((((((.....((.......))..(((((((((((........)).))))))))).))))))))...))........... (-25.60 = -26.86 + 1.26)

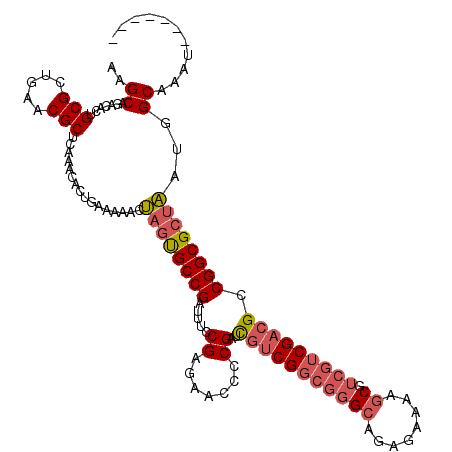
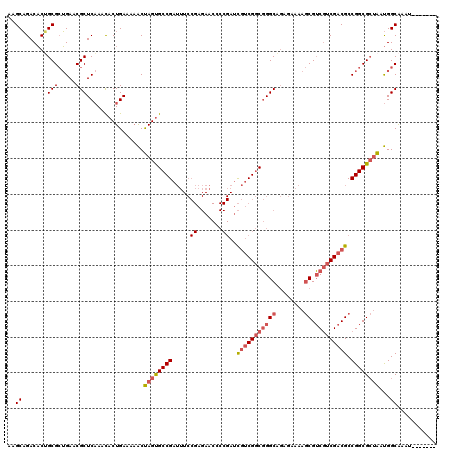
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:18 2006