
| Sequence ID | X_DroMel_CAF1 |
|---|---|
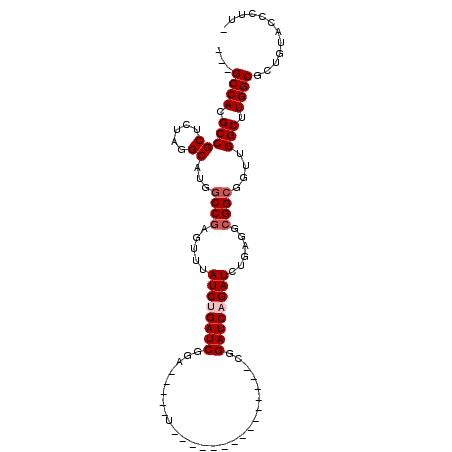
| Location | 19,318,110 – 19,318,217 |
| Length | 107 |
| Max. P | 0.990005 |

| Location | 19,318,110 – 19,318,217 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -24.57 |
| Energy contribution | -25.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
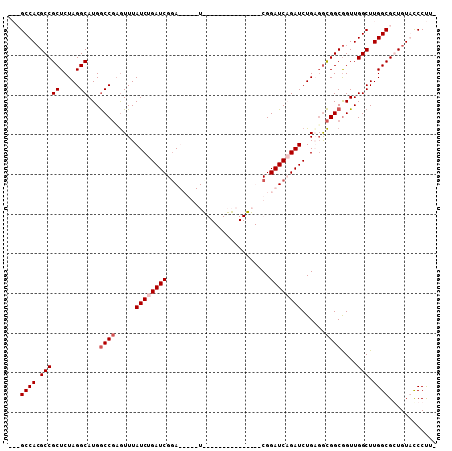
>X_DroMel_CAF1 19318110 107 + 22224390 GACGCCAAGCCGCUCUAGGCAUGGCCGAGUUUAUCUGAUCGGAUCGGAUCGGAUUGGACCGGUACGGAUCAGAUCUGAGGCGGCAGUUGGCUUGGCGCUGUGCCCUU- ..(((((((((((.....))...((((..((.((((((((.((((((.((......))))))).).))))))))..))..))))....)))))))))..........- ( -49.00) >DroSec_CAF1 8411 102 + 1 GACGCCACGCCGCUCUAGGCAUGGCCGCGUUUAUCUGAUCGGA-----UUGGAUUGGACCGGUACGGAUCAGAUCUGAGGCGGCGGUUGGCUUGGCGCUGUACCCUU- ..(((((.(((((.....))((.(((((.((.((((((((.((-----((((......))))).).))))))))..)).))))).)).))).)))))..........- ( -46.60) >DroEre_CAF1 10149 85 + 1 ---GCCACGCCGCUCUAGGCAUGACCUAAUUUAUCUGAUCGUA-----U---------------CGGAUCUGAUCUGUGGUGGCGGUUGGCUUGGCGGUGUAUCCCUU ---...((((((((..((((.((.((..((..(((.((((...-----.---------------..)))).)))..))...))))....))))))))))))....... ( -26.40) >DroYak_CAF1 9706 90 + 1 ---GCCACGCCGCUCUAGGCAUGGCCGAGUUUAUCUGAUCGGAUCGGAU---------------CGGAUCUGAUCUGUUACGGCGGUUGGCUUGGCGGUGUGCCCUUC ---..(((((((((..((((.((((((.((....(.(((((((((....---------------..))))))))).)..))..)))))))))))))))))))...... ( -39.70) >consensus ___GCCACGCCGCUCUAGGCAUGGCCGAGUUUAUCUGAUCGGA_____U_______________CGGAUCAGAUCUGAGGCGGCGGUUGGCUUGGCGCUGUACCCUU_ ...((((.(((((.....))...((((.....((((((((..........................))))))))......))))....))).))))............ (-24.57 = -25.57 + 1.00)

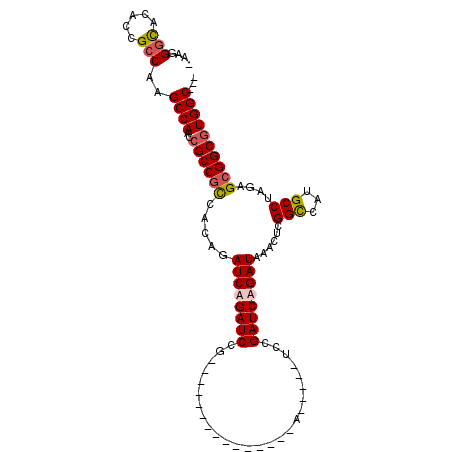
| Location | 19,318,110 – 19,318,217 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.96 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
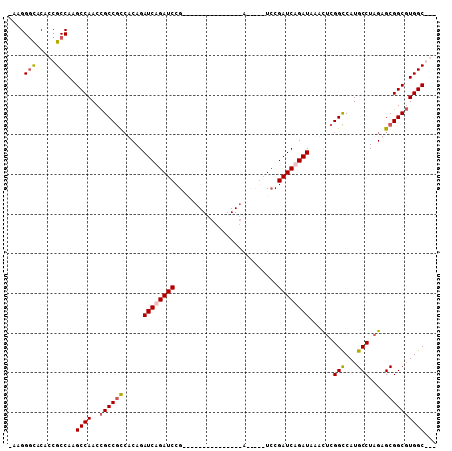
>X_DroMel_CAF1 19318110 107 - 22224390 -AAGGGCACAGCGCCAAGCCAACUGCCGCCUCAGAUCUGAUCCGUACCGGUCCAAUCCGAUCCGAUCCGAUCAGAUAAACUCGGCCAUGCCUAGAGCGGCUUGGCGUC -.........((((((((((....((((......((((((((.(...(((((......)).)))..).)))))))).....))))...((.....)))))))))))). ( -42.00) >DroSec_CAF1 8411 102 - 1 -AAGGGUACAGCGCCAAGCCAACCGCCGCCUCAGAUCUGAUCCGUACCGGUCCAAUCCAA-----UCCGAUCAGAUAAACGCGGCCAUGCCUAGAGCGGCGUGGCGUC -.........((((((.(((....(((((.....((((((((.(....((......))..-----.).))))))))....)))))...((.....))))).)))))). ( -38.80) >DroEre_CAF1 10149 85 - 1 AAGGGAUACACCGCCAAGCCAACCGCCACCACAGAUCAGAUCCG---------------A-----UACGAUCAGAUAAAUUAGGUCAUGCCUAGAGCGGCGUGGC--- ..((......)).....((((..((((.......(((.((((..---------------.-----...)))).)))...(((((.....)))))...))))))))--- ( -23.30) >DroYak_CAF1 9706 90 - 1 GAAGGGCACACCGCCAAGCCAACCGCCGUAACAGAUCAGAUCCG---------------AUCCGAUCCGAUCAGAUAAACUCGGCCAUGCCUAGAGCGGCGUGGC--- ....(((.....)))..((((..((((((....((((.((((..---------------....)))).)))).......((.(((...))).)).))))))))))--- ( -32.30) >consensus _AAGGGCACACCGCCAAGCCAACCGCCGCCACAGAUCAGAUCCG_______________A_____UCCGAUCAGAUAAACUCGGCCAUGCCUAGAGCGGCGUGGC___ ....(((.....)))..((((..((((((.....((((((((..........................))))))))......(((...)))....))))))))))... (-24.09 = -24.72 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:05 2006