
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,314,725 – 19,314,834 |
| Length | 109 |
| Max. P | 0.886402 |

| Location | 19,314,725 – 19,314,834 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -35.46 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
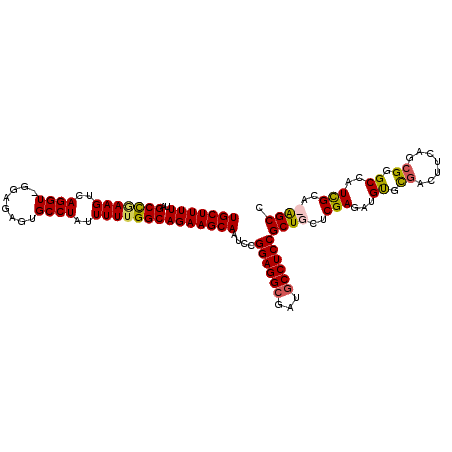
>X_DroMel_CAF1 19314725 109 + 22224390 UGCUUUUUAGCCGAAGUCAGGU-GGAGAGUGCCUAUUUUUGGCAGAAGCAAUCCGGAGGCGAUGCCUCCGCUGCUCGAGAUGUGUGGCUUCAGCGGGCGAUUGCACAGCU .(((.....(((((((..((((-.......))))..)))))))....(((((((((((((...)))))))(((((.(((........))).)))))..))))))..))). ( -43.90) >DroSec_CAF1 4770 109 + 1 UGCUUUUUAGCUGAAGUCAGGU-GUAGAGUGCCUAUUUUUGGCAGAAGCAAUCCGGAGGCGAUUCCUCCGCUGCUCGAGAUGUGCGGCUUCAGCGGGCAAUUGCACAGCU .(((.....(((((((((((((-.......))))(((((.(((((........((((((.....))))))))))).)))))....)))))))))..((....))..))). ( -39.20) >DroEre_CAF1 6764 107 + 1 UGCUUUUUCGCCAAAGUCAGGU-GAAGAGUGCCUAUUUUUGGCAGAAGCAAACCGGAGGCAAUGCCUCCGCUGCUCGAGAUGCGCGACUUCAG-GGGCCAUCGCA-GGCC (((((((..(((((((..((((-.......))))..))))))))))))))...(((((((...)))))))((((....((((.((..(....)-..)))))))))-)... ( -42.50) >DroYak_CAF1 6299 109 + 1 UGCUUUUUCGCCCAAGUCAGGUUGGAGAGUGCCUAUUUUUGGCAGAAGCAAUCCGGAGGCGAUGCCUCCGCUGCUCGAGAUGUGCGACAACGGCGGGCCAUCGCG-GGCC (((((((..(((.(((..((((........))))..))).))))))))))...(((((((...)))))))..(((((.((((.((..(......).)))))).))-))). ( -41.90) >consensus UGCUUUUUAGCCGAAGUCAGGU_GGAGAGUGCCUAUUUUUGGCAGAAGCAAUCCGGAGGCGAUGCCUCCGCUGCUCGAGAUGUGCGACUUCAGCGGGCCAUCGCA_AGCC (((((((..(((((((..((((........))))..))))))))))))))....((((((...))))))((((..(((...((.((.......)).))..)))..)))). (-35.46 = -35.52 + 0.06)

| Location | 19,314,725 – 19,314,834 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.57 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -31.94 |
| Energy contribution | -32.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19314725 109 - 22224390 AGCUGUGCAAUCGCCCGCUGAAGCCACACAUCUCGAGCAGCGGAGGCAUCGCCUCCGGAUUGCUUCUGCCAAAAAUAGGCACUCUCC-ACCUGACUUCGGCUAAAAAGCA .((((.((....)).)(((((((.((........(((((((((((((...)))))))..)))))).((((.......))))......-...)).))))))).....))). ( -36.60) >DroSec_CAF1 4770 109 - 1 AGCUGUGCAAUUGCCCGCUGAAGCCGCACAUCUCGAGCAGCGGAGGAAUCGCCUCCGGAUUGCUUCUGCCAAAAAUAGGCACUCUAC-ACCUGACUUCAGCUAAAAAGCA .((((.((....)).)(((((((.(.........((((((((((((.....))))))..)))))).((((.......))))......-....).))))))).....))). ( -35.10) >DroEre_CAF1 6764 107 - 1 GGCC-UGCGAUGGCCC-CUGAAGUCGCGCAUCUCGAGCAGCGGAGGCAUUGCCUCCGGUUUGCUUCUGCCAAAAAUAGGCACUCUUC-ACCUGACUUUGGCGAAAAAGCA ((((-......))))(-(..(((((.........(((((((((((((...)))))))..)))))).((((.......))))......-....)))))..).)........ ( -38.30) >DroYak_CAF1 6299 109 - 1 GGCC-CGCGAUGGCCCGCCGUUGUCGCACAUCUCGAGCAGCGGAGGCAUCGCCUCCGGAUUGCUUCUGCCAAAAAUAGGCACUCUCCAACCUGACUUGGGCGAAAAAGCA .(((-(((((..((.....))..)))).......((((((.((((((...))))))(((..(....((((.......)))))..)))...))).)))))))......... ( -41.00) >consensus AGCC_UGCAAUGGCCCGCUGAAGCCGCACAUCUCGAGCAGCGGAGGCAUCGCCUCCGGAUUGCUUCUGCCAAAAAUAGGCACUCUCC_ACCUGACUUCGGCGAAAAAGCA .((...((....))..((((((((((........((((((((((((.....))))))..)))))).((((.......))))..........))))))))))......)). (-31.94 = -32.75 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:47:04 2006