| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,305,166 – 19,305,273 |
| Length | 107 |
| Max. P | 0.999786 |

| Location | 19,305,166 – 19,305,273 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.82 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19305166 107 + 22224390 CUCAGGCUCUGCCAAACU-AAUUUAUUUUUGAGCUUGUGGGGAAGGGAGGAGCAGGUGGAAAUAGAGAACGACACUCUAAAUAAAAAUUAAAA-------AAGGGCAGGCCUGA-----A .((((((.(((((.....-.....(((((((.((((((.............)))))).....(((((.......)))))..))))))).....-------...)))))))))))-----. ( -30.79) >DroSec_CAF1 83559 91 + 1 CUCAGGCGCUGCCAAACUUAAAUUAUU-UCGAGC--------AAGGGAGGAGCGGGAGGAAAUAGAGGAGGACACUCCAAAUGGAA---------------UAGGGAGGCCUGA-----A .((((((.((.((.........(((((-((..((--------.........)).....))))))).((((....))))........---------------..)).))))))))-----. ( -27.10) >DroEre_CAF1 79603 97 + 1 CUCAGGCACUGCCAAACU-ACUUUAUU-UCGAGG------------GAUCAGC----AAAGAUCUCGGAAAAGACUCGAAAGAAAAUAUAGUUCGUUGCCCCAGGCAGGCCUGA-----G (((((((.(((((.....-......((-((..((------------((((...----...)))))).))))....((....))....................)))))))))))-----) ( -35.10) >DroYak_CAF1 84989 102 + 1 CUCAGGCACUGCCAAACU-ACUUCAUU-UCGAGC------------GUGUGGG----GGAAAUAGACGAGGACACUCUAAAGGAAAUGUGGCUUGUUGCCCCAGGCAGGCCUGAAAAAAG .((((((.(((((....(-(((((...-..))).------------)))..((----((.....((((((..(((............))).)))))).)))).)))))))))))...... ( -38.20) >consensus CUCAGGCACUGCCAAACU_AAUUUAUU_UCGAGC____________GAGGAGC____GGAAAUAGAGGAGGACACUCUAAAGAAAAU_UAG_U_______CCAGGCAGGCCUGA_____A .((((((.(((((.........((......))..............................(((((.......)))))........................)))))))))))...... (-17.24 = -18.05 + 0.81)

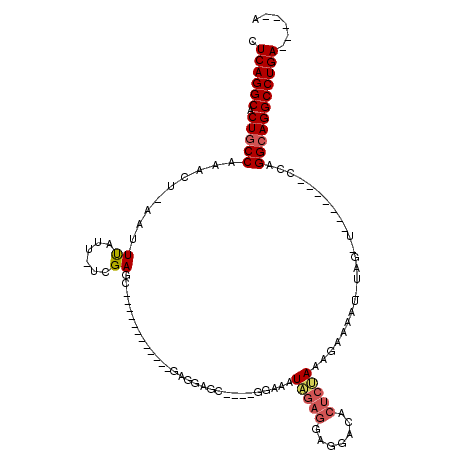
| Location | 19,305,166 – 19,305,273 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.82 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -17.20 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.56 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19305166 107 - 22224390 U-----UCAGGCCUGCCCUU-------UUUUAAUUUUUAUUUAGAGUGUCGUUCUCUAUUUCCACCUGCUCCUCCCUUCCCCACAAGCUCAAAAAUAAAUU-AGUUUGGCAGAGCCUGAG .-----(((((((((((((.-------.((((.(((((...(((((.......))))).........(((...............)))..)))))))))..-))...))))).)))))). ( -25.96) >DroSec_CAF1 83559 91 - 1 U-----UCAGGCCUCCCUA---------------UUCCAUUUGGAGUGUCCUCCUCUAUUUCCUCCCGCUCCUCCCUU--------GCUCGA-AAUAAUUUAAGUUUGGCAGCGCCUGAG .-----((((((.....((---------------((((....))))))...................(((....((..--------(((..(-(....))..)))..)).))))))))). ( -22.60) >DroEre_CAF1 79603 97 - 1 C-----UCAGGCCUGCCUGGGGCAACGAACUAUAUUUUCUUUCGAGUCUUUUCCGAGAUCUUU----GCUGAUC------------CCUCGA-AAUAAAGU-AGUUUGGCAGUGCCUGAG (-----(((((((((((...(....)(((((((..(((.(((((((((......))((((...----...))))------------.)))))-)).)))))-)))))))))).))))))) ( -39.20) >DroYak_CAF1 84989 102 - 1 CUUUUUUCAGGCCUGCCUGGGGCAACAAGCCACAUUUCCUUUAGAGUGUCCUCGUCUAUUUCC----CCCACAC------------GCUCGA-AAUGAAGU-AGUUUGGCAGUGCCUGAG ......(((((((((((...(((.....)))..(((((.(((.((((((..............----.....))------------)))).)-)).)))))-.....))))).)))))). ( -34.01) >consensus C_____UCAGGCCUGCCUGG_______A_CUA_AUUUCAUUUAGAGUGUCCUCCUCUAUUUCC____GCUCCUC____________GCUCGA_AAUAAAGU_AGUUUGGCAGUGCCUGAG ......(((((((((((...(....)...............(((((.......))))).................................................))))).)))))). (-17.20 = -17.82 + 0.62)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:56 2006