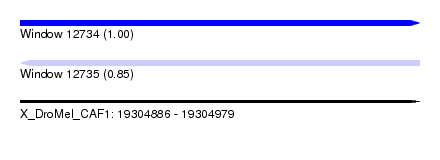
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,304,886 – 19,304,979 |
| Length | 93 |
| Max. P | 0.999866 |

| Location | 19,304,886 – 19,304,979 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 63.22 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.50 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
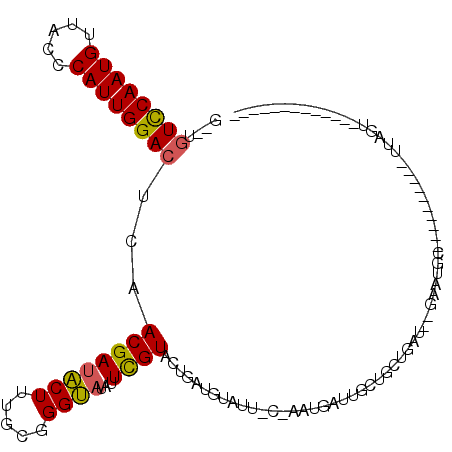
>X_DroMel_CAF1 19304886 93 + 22224390 -------------ACUAA----------GCAUCCAAAUCAGCAGCAAUCAUUCGCAAUACAUCAGUACGAAUAUACCCACAAAGUGUCGUUGAGUCCAAUGGGUAACAUUGAAUG--C -------------.....----------((((.(((.............(((((...(((....)))))))).(((((((.....)).((((....)))))))))...))).)))--) ( -14.60) >DroSec_CAF1 83270 81 + 1 -------------ACCAA----------GCAUUC---UCAUCAGCAAUC-----------AGCAGUACGAAUAUACCCGCAAAGUAUCGUUGAGUCCAAUGGGUAACAUUGGACAGCC -------------.....----------......---......((..((-----------(((.((((...............)))).)))))((((((((.....)))))))).)). ( -21.76) >DroEre_CAF1 79273 116 + 1 AAUAACGAAGCAAACGAAUGCAUAUUAGGCAUUC--AUCAGCCAUCAUCAUUUGAAAUACAUCAGAACAAAUAUGCCCGCAAAGCAUCGUUAAGUCCAAUGGGUAACAUUGGACAAAU ..((((((.((...((...(((((((.(((....--....))).......(((((......)))))...))))))).))....)).)))))).((((((((.....)))))))).... ( -34.10) >consensus _____________ACCAA__________GCAUUC__AUCAGCAGCAAUCAUU_G_AAUACAUCAGUACGAAUAUACCCGCAAAGUAUCGUUGAGUCCAAUGGGUAACAUUGGACA__C ...................................................................(((..((((.......))))..))).((((((((.....)))))))).... (-11.77 = -11.33 + -0.44)


| Location | 19,304,886 – 19,304,979 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 63.22 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -11.88 |
| Energy contribution | -11.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19304886 93 - 22224390 G--CAUUCAAUGUUACCCAUUGGACUCAACGACACUUUGUGGGUAUAUUCGUACUGAUGUAUUGCGAAUGAUUGCUGCUGAUUUGGAUGC----------UUAGU------------- (--(((((((...(((((((.((...........))..)))))))((((((((.........))))))))............))))))))----------.....------------- ( -22.80) >DroSec_CAF1 83270 81 - 1 GGCUGUCCAAUGUUACCCAUUGGACUCAACGAUACUUUGCGGGUAUAUUCGUACUGCU-----------GAUUGCUGAUGA---GAAUGC----------UUGGU------------- ....((((((((.....))))))))..............(((((((.(((((...((.-----------....))..))))---).))))----------)))..------------- ( -23.50) >DroEre_CAF1 79273 116 - 1 AUUUGUCCAAUGUUACCCAUUGGACUUAACGAUGCUUUGCGGGCAUAUUUGUUCUGAUGUAUUUCAAAUGAUGAUGGCUGAU--GAAUGCCUAAUAUGCAUUCGUUUGCUUCGUUAUU ....((((((((.....)))))))).....(((((.(((.(((((....)))))))).)))))...((((((((.(((.(((--((((((.......))))))))).))))))))))) ( -36.20) >consensus G__UGUCCAAUGUUACCCAUUGGACUCAACGAUACUUUGCGGGUAUAUUCGUACUGAUGUAUU_C_AAUGAUUGCUGCUGAU__GAAUGC__________UUAGU_____________ ....((((((((.....))))))))...((((((((.....))))...)))).................................................................. (-11.88 = -11.67 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:54 2006