
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,293,450 – 19,293,588 |
| Length | 138 |
| Max. P | 0.999612 |

| Location | 19,293,450 – 19,293,558 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -29.81 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19293450 108 - 22224390 ACUGAUGGCGACGAACGAUGCAAGAACACUUU-----CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAA-ACUUCUGU------GCACUCCUG ((..((((.(((...(((.((((((((((...-----.........)))))))))).))).....)))))))..))(((((((........)))))-))......------......... ( -29.60) >DroSec_CAF1 72083 113 - 1 ACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAA-ACUUCUGC------GCACUCCUG .....((((((.....((.((((((((((.................)))))))))).))(((((...((....))..)))))....))))))(((.-.....)))------......... ( -29.53) >DroSim_CAF1 76485 113 - 1 ACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAA-ACUUCUGC------GCACUCCUG .....((((((.....((.((((((((((.................)))))))))).))(((((...((....))..)))))....))))))(((.-.....)))------......... ( -29.53) >DroEre_CAF1 68152 112 - 1 ACUGAUGGCGACGAACGAUGCAAGAACACUUCU-------UCACAUGUGUUUUUGUUUCGCAA-AGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAAAACUUCUGCACUUCCGCACUCCUG ..((.((((((.....((.((((((((((....-------......)))))))))).))((((-(..((....))..)))))....)))))).)).......(((......)))...... ( -30.00) >DroYak_CAF1 73235 105 - 1 ACUGAUGGCGACGAACGAUGCAAGAACACUUCU-------UCACAUGUGUUUUUGUUUCGCAA-AGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAA-ACUUCUGC------GCACUCCUG ((..((((.(((...(((.((((((((((....-------......)))))))))).)))...-.)))))))..))(((((((........)))))-))......------......... ( -30.40) >consensus ACUGAUGGCGACGAACGAUGCAAGAACACUUCC____CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUUGCUCUUUCGCCAGCAA_ACUUCUGC______GCACUCCUG ..((.((((((.....((.((((((((((.................)))))))))).))(((((...((....))..)))))....)))))).)).......(((......)))...... (-27.03 = -27.27 + 0.24)

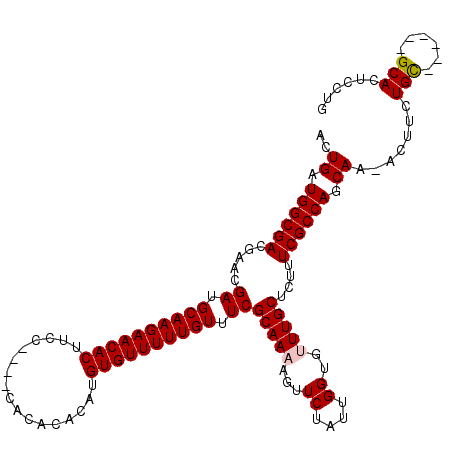
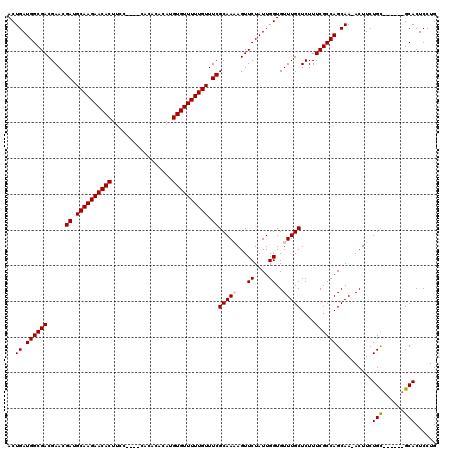
| Location | 19,293,483 – 19,293,588 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19293483 105 + 22224390 AAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUG-----AAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAA----------GCGACGAAAAGCUAAUAUGUGU ..((((...(((..(((((((((((((...((((....)))).-----....)))).)))).(((((..((((((....))))))..)----------))))..))))))))....)))) ( -30.30) >DroSec_CAF1 72116 110 + 1 AAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAA----------GCGACGAAAAGCUAAUAUGUGU ..((((...(((..(((((((((((((...(((((....)))))........)))).)))).(((((..((((((....))))))..)----------))))..))))))))....)))) ( -28.30) >DroSim_CAF1 76518 110 + 1 AAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUGUAAUGGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAA----------GCGACGAAAAGCUAAUAUGUGU ..((((...(((..(((((((((((((...(((((....)))))........)))).)))).(((((..((((((....))))))..)----------))))..))))))))....)))) ( -28.30) >DroEre_CAF1 68192 112 + 1 AAACACCAAUAGAACU-UUGCGAAACAAAAACACAUGUGA-------AGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAAGCGAGGAAAAGCGACGAAAAGCUAAUAUGUAU ..(((((.........-.(((((((((....((....)).-------.....)))).)))))(((((..((((((....))))))..)))))))...(((........)))....))).. ( -27.00) >DroYak_CAF1 73268 102 + 1 AAACACCAAUAGAACU-UUGCGAAACAAAAACACAUGUGA-------AGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAA----------GCGACGAAAAGCUAAUAUGUAU ..(((....(((..((-((.((...(((.(((((......-------....))))).)))..(((((..((((((....))))))..)----------)))))).)))))))...))).. ( -23.80) >consensus AAACACCAAUAGAACUUUUGCGAAACAAAAACACAUGUGUGUG____AGAAGUGUUCUUGCAUCGUUCGUCGCCAUCAGUGGCGAAAA__________GCGACGAAAAGCUAAUAUGUGU ..((((...(((.....((((((..(((.(((((.................))))).)))..)).((((((((.....))))))))............)))).......)))....)))) (-19.53 = -19.93 + 0.40)

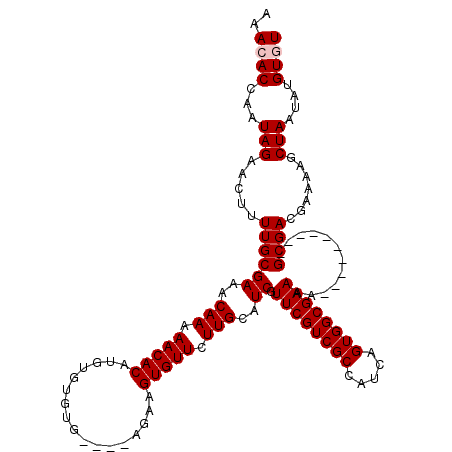
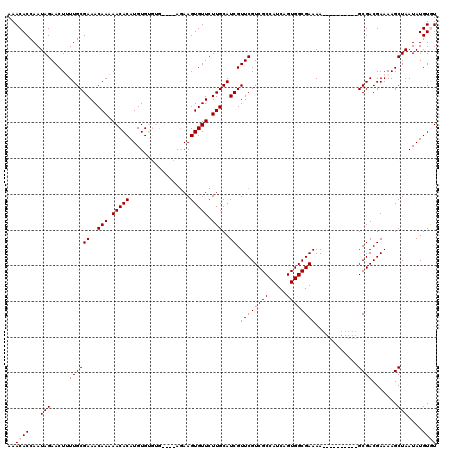
| Location | 19,293,483 – 19,293,588 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.04 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -26.99 |
| Energy contribution | -27.15 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19293483 105 - 22224390 ACACAUAUUAGCUUUUCGUCGC----------UUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUU-----CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUU ((((.....(((((((((((((----------(..(((....))).)))))))).(((.((((((((((...-----.........)))))))))).)))..))))))......)))).. ( -33.30) >DroSec_CAF1 72116 110 - 1 ACACAUAUUAGCUUUUCGUCGC----------UUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUU ((((.....(((((((((((((----------(..(((....))).)))))))).(((.((((((((((.................)))))))))).)))..))))))......)))).. ( -32.93) >DroSim_CAF1 76518 110 - 1 ACACAUAUUAGCUUUUCGUCGC----------UUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCCAUUACACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUU ((((.....(((((((((((((----------(..(((....))).)))))))).(((.((((((((((.................)))))))))).)))..))))))......)))).. ( -32.93) >DroEre_CAF1 68192 112 - 1 AUACAUAUUAGCUUUUCGUCGCUUUUCCUCGCUUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCU-------UCACAUGUGUUUUUGUUUCGCAA-AGUUCUAUUGGUGUUU ((((.....((((((((((((((.......((.....)).......)))))))).(((.((((((((((....-------......)))))))))).))).))-))))......)))).. ( -32.24) >DroYak_CAF1 73268 102 - 1 AUACAUAUUAGCUUUUCGUCGC----------UUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCU-------UCACAUGUGUUUUUGUUUCGCAA-AGUUCUAUUGGUGUUU ((((.....(((((((((((((----------(..(((....))).)))))))).(((.((((((((((....-------......)))))))))).))).))-))))......)))).. ( -31.60) >consensus ACACAUAUUAGCUUUUCGUCGC__________UUUUCGCCACUGAUGGCGACGAACGAUGCAAGAACACUUCC____CACACACAUGUGUUUUUGUUUCGCAAAAGUUCUAUUGGUGUUU ((((.....(((((((...................((((((....))))))....(((.((((((((((.................)))))))))).))).)))))))......)))).. (-26.99 = -27.15 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:49 2006