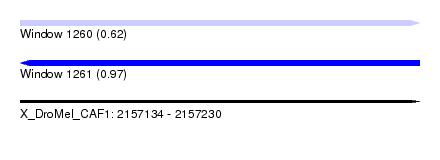
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,157,134 – 2,157,230 |
| Length | 96 |
| Max. P | 0.973079 |

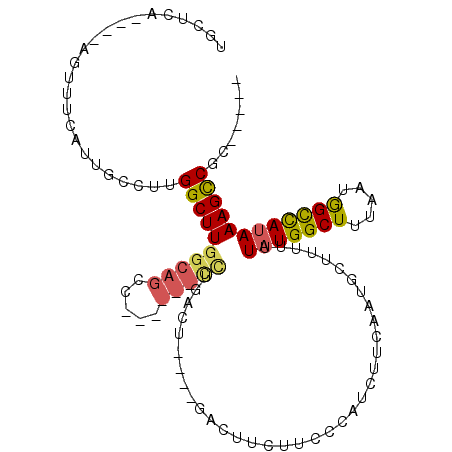
| Location | 2,157,134 – 2,157,230 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -9.14 |
| Energy contribution | -9.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2157134 96 + 22224390 -----GCGGCUUUGUAGCUAUAAAAGCCAUAAAAAAGCGUUGAAGAUGCCAAGAAGUC----ACUCAAA------GCCAGCAAAGCCAAGAAAAUGAAACGGCCAGGCGGA -----..(((((((..(((.....))).........(((((...))))).........----...))))------))).((...(((.............)))...))... ( -20.32) >DroPse_CAF1 50116 98 + 1 -----GCGGCUUUAUGGCCAUUAAAGCCAUAAAAAAGCAUUGAAGAUGGGAAAAAGUC----AGUCAGGCAGGCUGGCUGCCAAGCCAAGGCAAUGAAAAU----UGAGCA -----(((((((((((((.......))))))))...............((....((((----((((.....)))))))).))..)))....((((....))----)).)). ( -32.50) >DroAna_CAF1 44450 93 + 1 CGGCAUCGACUUUAUGACCAUAAAAGCCAUAAAAAAGCAUUGAAGAUGGCAAGAAGUCAGUCACUCAAA------GC----CAAGCCAAGAAAAUUGAACGG--------A .(((...((((((.((.((((.((.((.........)).))....)))))).)))))).((........------))----...)))...............--------. ( -16.80) >DroPer_CAF1 51023 98 + 1 -----GCGGCUUUAUGGCCAUUAAAGCCAUAAAAAAGCAUUGAAGAUGGGAAAAAGUC----AGUCAGGCAGGCUGGCUGCCAAGCCAAGGCAAUGAAAAU----UGAGCA -----(((((((((((((.......))))))))...............((....((((----((((.....)))))))).))..)))....((((....))----)).)). ( -32.50) >consensus _____GCGGCUUUAUGGCCAUAAAAGCCAUAAAAAAGCAUUGAAGAUGGCAAAAAGUC____ACUCAAA______GCCUGCCAAGCCAAGAAAAUGAAAAG____UGAGCA .......(((((((((((.......))))))))......((((.(((........)))......))))................)))........................ ( -9.14 = -9.45 + 0.31)

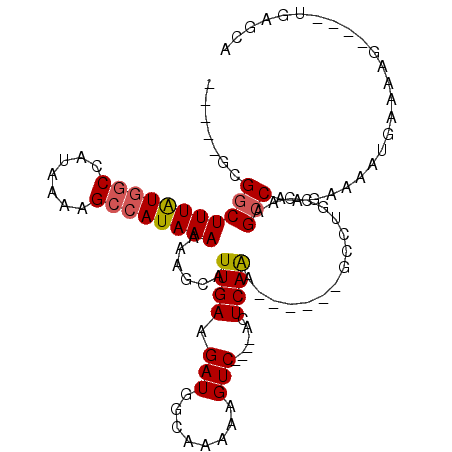
| Location | 2,157,134 – 2,157,230 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -13.56 |
| Energy contribution | -14.25 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.973079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2157134 96 - 22224390 UCCGCCUGGCCGUUUCAUUUUCUUGGCUUUGCUGGC------UUUGAGU----GACUUCUUGGCAUCUUCAACGCUUUUUUAUGGCUUUUAUAGCUACAAAGCCGC----- ...((..(((((...........)))))..)).(((------(((((((----(.....((((.....))))))))......(((((.....))))))))))))..----- ( -24.50) >DroPse_CAF1 50116 98 - 1 UGCUCA----AUUUUCAUUGCCUUGGCUUGGCAGCCAGCCUGCCUGACU----GACUUUUUCCCAUCUUCAAUGCUUUUUUAUGGCUUUAAUGGCCAUAAAGCCGC----- ......----..............((((((((((.....))))).....----...........................(((((((.....))))))))))))..----- ( -23.50) >DroAna_CAF1 44450 93 - 1 U--------CCGUUCAAUUUUCUUGGCUUG----GC------UUUGAGUGACUGACUUCUUGCCAUCUUCAAUGCUUUUUUAUGGCUUUUAUGGUCAUAAAGUCGAUGCCG .--------...............((((((----((------(((..((((((........(((((...............)))))......)))))))))))))).))). ( -22.80) >DroPer_CAF1 51023 98 - 1 UGCUCA----AUUUUCAUUGCCUUGGCUUGGCAGCCAGCCUGCCUGACU----GACUUUUUCCCAUCUUCAAUGCUUUUUUAUGGCUUUAAUGGCCAUAAAGCCGC----- ......----..............((((((((((.....))))).....----...........................(((((((.....))))))))))))..----- ( -23.50) >consensus UGCUCA____AGUUUCAUUGCCUUGGCUUGGCAGCC______CCUGACU____GACUUCUUCCCAUCUUCAAUGCUUUUUUAUGGCUUUAAUGGCCAUAAAGCCGC_____ ........................((((((((((.....)))))....................................(((((((.....))))))))))))....... (-13.56 = -14.25 + 0.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:34 2006