
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,279,385 – 19,279,505 |
| Length | 120 |
| Max. P | 0.588429 |

| Location | 19,279,385 – 19,279,486 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -19.86 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19279385 101 + 22224390 ------------------CACCAUCCAUGCUUUCACCUACGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCAGGCCUCCGGGCAUUGGGCGAACGAAAUCUUUAUGGAUUUUGCCAUAU ------------------.....................((((((((((....)).............(((.((...)).)))))))))))..((((((((....))))))))...... ( -23.50) >DroSec_CAF1 58649 114 + 1 -----CCACCCAGUUCCCCACUUUCCACGCCCCCACCACCGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCGGGCCUGCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGU -----.......................((((.((.....((((...........................)))).)).))))....(((((((((........)))..)))))).... ( -23.73) >DroSim_CAF1 62943 114 + 1 -----CCACCCAGUUCCCCACUUUCCACGCCCCCACCGCCGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCGGGCCUGCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGU -----......................(((((...((((.((((...........................))))..)))).....)))))...........(((((......))))). ( -26.33) >DroEre_CAF1 55265 105 + 1 --------------UCCCCACUUUCCACGCUUUCACCUCCGCCCAAUCAUUUUUGCACCUAUAUUCCAUGAAGGCCUCCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUUGCCAUGU --------------........................................(((.......((((((((((((....))).((((.....))))..)))))))))...)))..... ( -20.40) >DroYak_CAF1 59439 118 + 1 AUCUCCCAAAC-UUCCCCCACUUUCCACGCUUUCACCUCCGCCCAAUCAUUUUUGCACCUAUAUUCGAGUUAGGCCUCCGGGCAUUGGGCGAACGAAAUCUUUAUGGAUUUCGCCAUGU ...........-................((.........((((((((...........(((.........)))(((....)))))))))))...(((((((....)))))))))..... ( -25.90) >consensus _____CCA__C__UUCCCCACUUUCCACGCUUUCACCUCCGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCAGGCCUCCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGU .......................................((((((((((....))..................(((....)))))))))))...........(((((......))))). (-19.86 = -19.70 + -0.16)

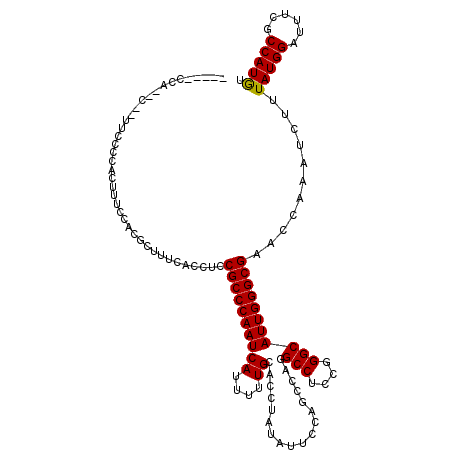
| Location | 19,279,406 – 19,279,505 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.13 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19279406 99 + 22224390 CGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCAGGCCUCCGGGCAUUGGGCGAACGAAAUCUUUAUGGAUUUUGCCAUAUAGUUGGAUUGACAUCCACU--------------------- ((((((((((....)).............(((.((...)).)))))))))))..((((((((....)))))))).........(((((....)))))..--------------------- ( -26.60) >DroSec_CAF1 58683 99 + 1 CGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCGGGCCUGCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGUAGUUGGAUUGACAUCCUCU--------------------- ((((((((..................((....))(((....)))))))))))..((((..((.(((((......))))).)))))).............--------------------- ( -26.50) >DroSim_CAF1 62977 99 + 1 CGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCGGGCCUGCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGUAGUUGGAUUGACAUCCUCU--------------------- ((((((((..................((....))(((....)))))))))))..((((..((.(((((......))))).)))))).............--------------------- ( -26.50) >DroEre_CAF1 55290 113 + 1 CGCCCAAUCAUUUUUGCACCUAUAUUCCAUGAAGGCCUCCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUUGCCAUGUAGUUGGAUUGACAUCCUCUGCUCCUCCUCCCUC------- ....(((((....(((((.......((((((((((((....))).((((.....))))..)))))))))........)))))...))))).......................------- ( -27.66) >DroYak_CAF1 59477 120 + 1 CGCCCAAUCAUUUUUGCACCUAUAUUCGAGUUAGGCCUCCGGGCAUUGGGCGAACGAAAUCUUUAUGGAUUUCGCCAUGUAGUUGGAUUGACAUCCUCUGCUCCUCCUCCCUCCCUCCCC ((((((((...........(((.........)))(((....)))))))))))..((((((((....))))))))....((((..((((....)))).))))................... ( -32.40) >consensus CGCCCAAUCAUUUUUGCACCUAUAUUCCAGCCAGGCCUCCGGGCAUUGGGCGAACCAAAUCUUUAUGGAUUUCGCCAUGUAGUUGGAUUGACAUCCUCU_____________________ ((((((((((....))..................(((....)))))))))))..............((((.((.(((......)))...)).))))........................ (-23.42 = -23.18 + -0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:43 2006