
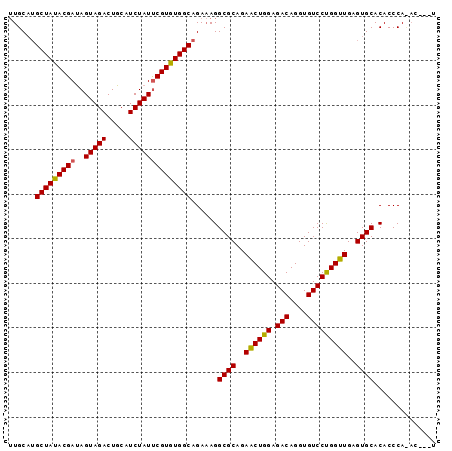
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,270,784 – 19,271,059 |
| Length | 275 |
| Max. P | 0.990595 |

| Location | 19,270,784 – 19,270,874 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -31.53 |
| Energy contribution | -31.20 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19270784 90 + 22224390 UUGCUUGCUAUACGAUAGUAGACUGCAUCUAUUCGUGUGGCAGAAAGGCGCAGAACUGGAGACAGGUGUCCCGGUUGAGUGCACACCCA------U ....(((((((((((..(((((.....))))))))))))))))....((((..((((((.(((....)))))))))..)))).......------. ( -34.70) >DroSim_CAF1 61823 96 + 1 UUGCAUGCUAUACGAUAGUAGACUGCAUCUAUUCGUGUGGCAGAAAGGCGCAGAACUGGAGACAGGUGUCCUGGUUGAGUGCACACCCAUACCCAU .....((((((((((..(((((.....))))))))))))))).....((((..(((..(.(((....))))..)))..)))).............. ( -31.70) >DroEre_CAF1 54191 91 + 1 AUGCAGGCUACACGACAGUAGACUGCAUCUAUCCGUGUGGCAGA-AGGCGCAGAGCUGGAGACAGGUGUCCUGGUUGAGUGCACACCCACAC---- ((((((.((((......)))).))))))......((((((....-..((((..(((..(.(((....))))..)))..))))....))))))---- ( -35.00) >consensus UUGCAUGCUAUACGAUAGUAGACUGCAUCUAUUCGUGUGGCAGAAAGGCGCAGAACUGGAGACAGGUGUCCUGGUUGAGUGCACACCCA_AC___U ......(((((((((..(((((.....))))))))))))))......((((..((((((.(((....)))))))))..)))).............. (-31.53 = -31.20 + -0.33)


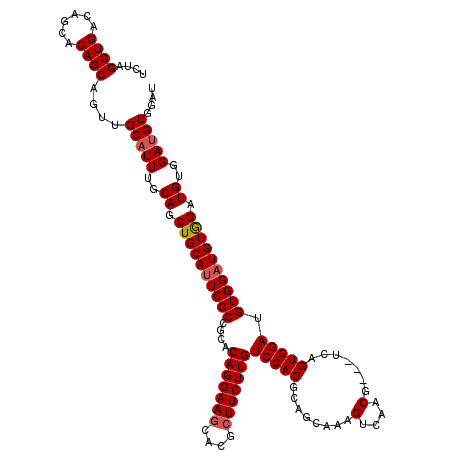
| Location | 19,270,874 – 19,270,991 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -37.70 |
| Energy contribution | -38.26 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19270874 117 - 22224390 UCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUCAACG---UUAGUGGAUGUGGAUGUAGAUGUGGAUGUGGAU ....((((......))))..(..((((..((.((((((((((....(((((((....)))))))(((((..(((.........)---)).))))).)))))))))).))..))))..).. ( -45.90) >DroSim_CAF1 61919 117 - 1 UCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUCAACG---UCAGUGGAUGUGGAUGUGGAUGUGGAAGUGGAU ....((.....))...((((((((....)))))))).....((((.(((((((....)))))))((((((...(...(((.(((---((....))))).)))...).)))))).)))).. ( -44.60) >DroEre_CAF1 54282 114 - 1 UCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCACUCGCCGCACAGGAAUCACGCUUCUUGUCCACGCAGCAAAGUCAACG---UCAGUGGAUGUGGAUGUGGAUGUGGAUGUG--- ............((((((((((((....)))))))).....(((((((((((......))))))(((((........(((.(((---((....))))).))))))))))))).))))--- ( -38.30) >DroYak_CAF1 58365 120 - 1 UCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUCAACGUCGUCAGUGGAUGUGGAUGUGGAUGAGGAUGUGGAU ............((((((((((((....)))))))).((((((((((((((((....)))))))((((((...((..(.(......).)..))..))))))))))).))))..))))... ( -41.80) >consensus UCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUCAACG___UCAGUGGAUGUGGAUGUGGAUGUGGAUGUGGAU ....((((......))))....(((((..((.((((((((((....(((((((....)))))))(((((........(....).......))))).)))))))))).))..))))).... (-37.70 = -38.26 + 0.56)


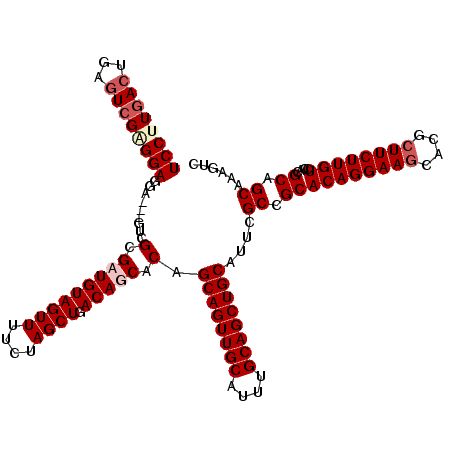
| Location | 19,270,911 – 19,271,028 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.48 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19270911 117 + 22224390 GACUUUGCUGCGUGGACAAGAAGCGUGCUUCCUGUGCGGCGAAUGCAGCUGCAAAUGCAACUGCUGUGCUGUCAGCUAGAAAACUACAACGCGAC---UCCUCCGCGAUUCAGUCAAGGA ..((((((((((((((..((..((((.((..(((.((((((...((((.(((....))).))))..)))))))))..)).........))))..)---)..))))))...))).))))). ( -45.00) >DroSim_CAF1 61956 117 + 1 GACUUUGCUGCGUGGACAAGAAGCGUGCUUCCUGUGCGGCGAAUGCAGCUGCAAAUGCAACUGCUGUGCUGUCAGCUAGAAAACUACAACGCGAC---UCCUCCUCGACUCGGUGAAGGA ..(((..(((((.(((..((..((((.((..(((.((((((...((((.(((....))).))))..)))))))))..)).........))))..)---)..))).))...)))..))).. ( -42.50) >DroEre_CAF1 54316 117 + 1 GACUUUGCUGCGUGGACAAGAAGCGUGAUUCCUGUGCGGCGAGUGCAGCUGCAAAUGCAACUGCUGUGCUGUCAGCUAGAAAACUACAUCGCGAC---UUCUCCCCGACUCAGUCAAGGA ..((((((((.((((...(((((((((((..(((.((((((...((((.(((....))).))))..))))))))).(((....))).)))))).)---))))..)).)).))).))))). ( -43.50) >DroYak_CAF1 58405 120 + 1 GACUUUGCUGCGUGGACAAGAAGCGUGCUUCCUGUGCGGCGAAUGCAGCUGCAAAUGCAACUGCUGUGCUGUCAGCUAGAAAACUACAUCGCGAGGAGGAGUCCUCGACUCAGUCAGGGA ..((((((((.((((....((((....))))(((.((((((...((((.(((....))).))))..)))))))))........))))....((((((....))))))...))).))))). ( -43.00) >consensus GACUUUGCUGCGUGGACAAGAAGCGUGCUUCCUGUGCGGCGAAUGCAGCUGCAAAUGCAACUGCUGUGCUGUCAGCUAGAAAACUACAACGCGAC___UCCUCCUCGACUCAGUCAAGGA ..(((..(((((.(((.......((((....(((.((((((...((((.(((....))).))))..))))))))).(((....)))...))))........))).))...)))..))).. (-33.86 = -33.48 + -0.38)



| Location | 19,270,911 – 19,271,028 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.97 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -37.24 |
| Energy contribution | -38.92 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19270911 117 - 22224390 UCCUUGACUGAAUCGCGGAGGA---GUCGCGUUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUC ..(((..(((....((((.(((---...(.(((((((((....)))).))))).).((((((((....)))))))).))).))))((((((((....)))))))).....)))..))).. ( -45.10) >DroSim_CAF1 61956 117 - 1 UCCUUCACCGAGUCGAGGAGGA---GUCGCGUUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUC (((((.((...)).)))))(((---((.(((((((((((....)))).)))))...((((((((....))))))))....)).)).(((((((....))))))))))............. ( -41.50) >DroEre_CAF1 54316 117 - 1 UCCUUGACUGAGUCGGGGAGAA---GUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCACUCGCCGCACAGGAAUCACGCUUCUUGUCCACGCAGCAAAGUC ..(((..(((.((.((.(((..---((.((....(((((....)))))...)))).((((((((....)))))))).))).))(.(((((((......))))))).))).)))..))).. ( -39.80) >DroYak_CAF1 58405 120 - 1 UCCCUGACUGAGUCGAGGACUCCUCCUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUC .....((((((((.(((((....)))))((....(((((....)))))...))...((((((((....))))))))))))((.((((((((((....))))))))....)).))..)))) ( -43.80) >consensus UCCUUGACUGAGUCGAGGAGGA___GUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUCGCCGCACAGGAAGCACGCUUCUUGUCCACGCAGCAAAGUC ((((((((...)))))))).........(.(((((((((....)))).))))).).((((((((....))))))))....((.((((((((((....))))))))....)).))...... (-37.24 = -38.92 + 1.69)


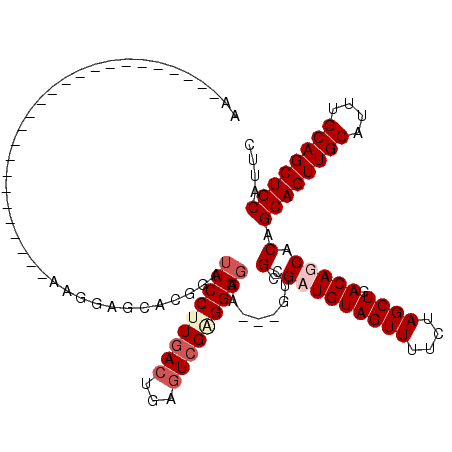
| Location | 19,270,951 – 19,271,059 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.15 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -25.36 |
| Energy contribution | -27.05 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19270951 108 - 22224390 AAGGAGCCA---------AAGGAGCCAAAGGAGCACCGGUUCCUUGACUGAAUCGCGGAGGA---GUCGCGUUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUC ((((((((.---------..((.(((....).)).))))))))))(((((...(((((....---.)))))...))))).....((.....))...((((((((....)))))))).... ( -39.30) >DroSim_CAF1 61996 90 - 1 ---------------------------AAGGAGCACUAGUUCCUUCACCGAGUCGAGGAGGA---GUCGCGUUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUC ---------------------------(((((((....)))))))...(((.((......))---.))).(((((((((....)))).)))))...((((((((....)))))))).... ( -33.40) >DroEre_CAF1 54356 90 - 1 AA---------------------------GGAGUACGGAUUCCUUGACUGAGUCGGGGAGAA---GUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCACUC ..---------------------------.((((.....(((((((((...)))))))))..---((.((....(((((....)))))...)))).((((((((....)))))))))))) ( -32.30) >DroYak_CAF1 58445 120 - 1 AAGGAGCCAGAGGAGCAAAAGGAGUCAAAGGAGCACGGAUUCCCUGACUGAGUCGAGGACUCCUCCUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUC .........((((((.....((((((...(.....).))))))......(((((...)))))))))))((....(((((....)))))...))...((((((((....)))))))).... ( -41.80) >consensus AA_________________________AAGGAGCACGGAUUCCUUGACUGAGUCGAGGAGGA___GUCGCGAUGUAGUUUUCUAGCUGACAGCACAGCAGUUGCAUUUGCAGCUGCAUUC .......................................(((((((((...)))))))))........(.(((((((((....)))).))))).).((((((((....)))))))).... (-25.36 = -27.05 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:41 2006