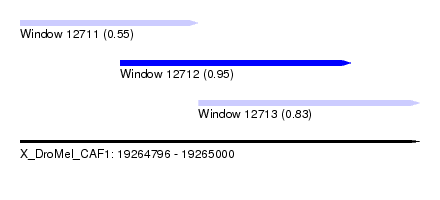
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,264,796 – 19,265,000 |
| Length | 204 |
| Max. P | 0.951104 |

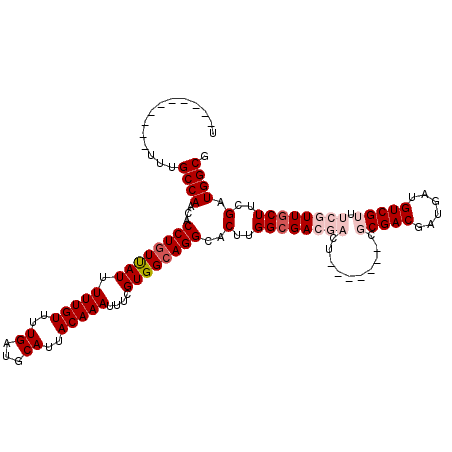
| Location | 19,264,796 – 19,264,887 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.17 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -8.54 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19264796 91 + 22224390 CCCUAUUAGAGGAUACUUGGCAACACUCAUUCCGUUUCCAC----------CU-------------------GCCCCUUUCCCCUUUGGCACACCUUUCGCUUAAGUGCAAUGUUGCAUU .(((.....))).......((((((................----------.(-------------------(((............))))........((......))..))))))... ( -14.90) >DroSec_CAF1 51882 91 + 1 CCCCAUUAGAGGGCACUUGGAAUCACUCAUUCCGUUUCCAC----------CU-------------------GCCCCUUGCCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUU .......((.(((((..(((((.............))))).----------.)-------------------))))))((((.....))))((((((.(((....))).)).)))).... ( -22.72) >DroSim_CAF1 56147 91 + 1 CCCCAUUGGAGGGCACUUGGAAUCACUCAUUCCGUUUCCAC----------CU-------------------GCCCCUUGCCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUU .......((((((((..(((((.............))))).----------.)-------------------))))..((((.....)))).....))).....((((((....)))))) ( -25.52) >DroEre_CAF1 48589 115 + 1 CCCUAUUAGAGGGUCCUUCGAAUCACUCAUUCCGUUUGCGCCCC-----ACCUUCUCGUCCUCGUUCCUCUCGCCCUCUUCCCUUUUGGCACAACUUGCACUUGAGUGCAAUGUUGCAUU .......(((((((.....((((.....))))((...(((....-----.......)))...))........)))))))..((....))..((((((((((....)))))).)))).... ( -22.50) >DroYak_CAF1 52382 100 + 1 CCCUAUUAGAGGGUCCUUGGAAUCACUCAUUCUGUUUGCGCUACGUAAAACCUC---A-----------------GCUUUCCCCCGUGGCACACCUUGCGCCUAAGUGCAGUGUUGCAUU ........((((......(((((.....))))).((((((...)))))).))))---.-----------------..........(..((((....(((((....)))))))))..)... ( -24.20) >consensus CCCUAUUAGAGGGUACUUGGAAUCACUCAUUCCGUUUCCAC__________CU___________________GCCCCUUUCCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUU .....((((((((.....(((((.....)))))(....)..........................................))))))))...............((((((....)))))) ( -8.54 = -9.30 + 0.76)

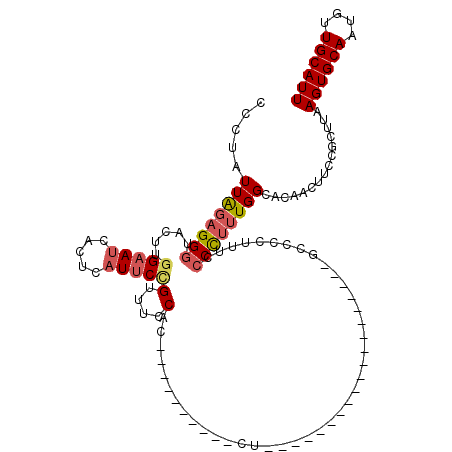
| Location | 19,264,847 – 19,264,965 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.25 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.44 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19264847 118 + 22224390 CCCCUUUGGCACACCUUUCGCUUAAGUGCAAUGUUGCAUUUUUUUUUUGUUUUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-- ...(((.(((.........))).)))((((....))))..........(((.(((((((..((((((((.(((((..((...))..)))))....))))))))...))))))).))).-- ( -28.40) >DroSec_CAF1 51933 108 + 1 CCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUUU----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGAGU-- .............((((.((((.((((((..(((((((...----------..)).))))).(((((((.(((((..((...))..)))))....)))))))))))))))))..))))-- ( -29.00) >DroSim_CAF1 56198 108 + 1 CCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUUU----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-- ..................((((.((((((..(((((((...----------..)).))))).(((((((.(((((..((...))..)))))....)))))))))))))))))......-- ( -28.00) >DroEre_CAF1 48664 108 + 1 CCCUUUUGGCACAACUUGCACUUGAGUGCAAUGUUGCAUUU----------UUGGCCAACACCUCUCAUUUUUGUUGUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-- .((....))..((((((((((....)))))).)))).....----------...(((((..(((.((((.....((((((....)))))).....)))).)))...))))).......-- ( -28.10) >DroYak_CAF1 52442 110 + 1 CCCCCGUGGCACACCUUGCGCCUAAGUGCAGUGUUGCAUUU----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACUCG ....(((.(((.....)))(((.((((((.((((((((...----------..)).))))))(((((((.(((((..((...))..)))))....)))))))))))))))).)))..... ( -35.20) >consensus CCCCUUUGGCACAACUUCCGCUUAAGUGCAAUGUUGCAUUU__________UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU__ ..................((((.((((((..(((((((...............)).))))).(((((((.(((((..((...))..)))))....)))))))))))))))))........ (-24.32 = -24.44 + 0.12)


| Location | 19,264,887 – 19,265,000 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -24.44 |
| Energy contribution | -26.28 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19264887 113 + 22224390 UUUUUUUUGUUUUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-------CGCGACGAUGAUGUCGUUUCGUUGCUUCGAUGGCG ..............((((...((((((((.(((((..((...))..)))))....))))))))..(..((((((((..-------.(((((......))))).))))))))..).)))). ( -34.70) >DroSec_CAF1 51973 103 + 1 U----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGAGU-------CGCGACGAUGAUGUCGUUUCGUUGCUUCGAUGGCG .----------...((((...((((((((.(((((..((...))..)))))....))))))))..(..(((((((((.-------.(((((......))))))))))))))..).)))). ( -35.60) >DroSim_CAF1 56238 103 + 1 U----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-------CGCGACGAUGAUGUCGUUUCGUUGCUUCGAUGGCG .----------...((((...((((((((.(((((..((...))..)))))....))))))))..(..((((((((..-------.(((((......))))).))))))))..).)))). ( -34.70) >DroEre_CAF1 48704 98 + 1 U----------UUGGCCAACACCUCUCAUUUUUGUUGUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU-------CGCGACGAUGAUGUCG-----UUGCUUCGAUGGCG .----------...((((...(((.((((.....((((((....)))))).....)))).)))..(..((((((((((-------((......))).))))-----)))))..).)))). ( -30.10) >DroYak_CAF1 52482 105 + 1 U----------UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACUCGCGACUCGCGACGAUGAUGUCG-----UUACUUCGAUGGCG .----------...((((.(.((((((((.(((((..((...))..)))))....))))))))....((((((((.((((((.....)).))).).)))))-----)))....).)))). ( -30.60) >consensus U__________UUUGCCAACACCUGUUAUUUUUGUUUUGAUGCAUUACAAAUUUCGUGGCAGGCACUUGGCGACGACU_______CGCGACGAUGAUGUCGUUUCGUUGCUUCGAUGGCG ..............((((...((((((((.(((((..((...))..)))))....))))))))..(..((((((((..........(((((......))))).))))))))..).)))). (-24.44 = -26.28 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:35 2006