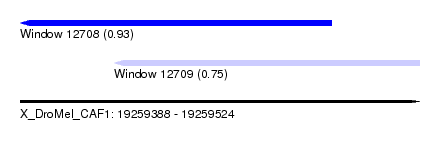
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,259,388 – 19,259,524 |
| Length | 136 |
| Max. P | 0.931921 |

| Location | 19,259,388 – 19,259,494 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -29.69 |
| Energy contribution | -30.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19259388 106 - 22224390 GUUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAGCAGC------UGAAACAGGCAACAGACGAAAGGAGCGA (((((((((((((.(((....)))(((((....)))))....)))))(((.((((....)))).(((((.......))------))).....))).......)))))))).. ( -35.10) >DroSec_CAF1 46322 106 - 1 GCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAGCAGC------UGAAACAGGCAACAGACGAAAGGAGCGA (((((((((((((.(((....)))(((((....)))))....)))))(((.((((....)))).(((((.......))------))).....))).......)))))))).. ( -37.50) >DroSim_CAF1 47618 106 - 1 GCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAGCAGC------UGAAACAGGCAACAGACGAAAGGAGCGA (((((((((((((.(((....)))(((((....)))))....)))))(((.((((....)))).(((((.......))------))).....))).......)))))))).. ( -37.50) >DroEre_CAF1 43257 104 - 1 GCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAGCAGCUGCAGCUGA--------ACAGAGGACAGGAGCGG (((((..(((..(((((....)))).)..)))..((((.((.(((((((((((((.(....((....))....)))))))))).)))--------).))..))))))))).. ( -40.80) >consensus GCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAGCAGC______UGAAACAGGCAACAGACGAAAGGAGCGA ((((((((((..(((((....)))).)..))).(((((...(((((..((.((((....))))....((....)).))......)))))..(....).)))))))))))).. (-29.69 = -30.75 + 1.06)


| Location | 19,259,420 – 19,259,524 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.46 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.10 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19259420 104 - 22224390 CAUCGCAAU---GCCCAUGCCCAUGCUCAUCCAGUUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAG ....(((..---((((((......(((.....)))......)))))).((((.....(((((....)))))...))))..))).((((....))))........... ( -26.90) >DroSec_CAF1 46354 92 - 1 ---------------CAUGCCCAUGCUCACCCAGCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAG ---------------.((((..(((((...((((((.(...(((((.(((....)))(((((....)))))....))))).).))))))....)))))..))))... ( -26.00) >DroSim_CAF1 47650 104 - 1 CAAGCCCAA---GCCCAUGCCCAUGCUCAUCCAGCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAG ...((....---((((((......(((.....)))......)))))).((((.....(((((....)))))...))))...)).((((....))))........... ( -26.90) >DroEre_CAF1 43287 98 - 1 ---------CAUCACCAUGCCCAUGCCCAUCCAGCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAG ---------.......((((..((((....((((((.(...(((((.(((....)))(((((....)))))....))))).).)))))).....))))..))))... ( -24.40) >consensus ____________GCCCAUGCCCAUGCUCAUCCAGCUCCUUUGUGGGCCGAAAUAUCGGACAAGCACUUGUCACUUUUCACUGCAGUUGGCAACAGCAUCAGCAUCAG ................((((..(((((...((((((.(...(((((.(((....)))(((((....)))))....))))).).))))))....)))))..))))... (-24.04 = -24.10 + 0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:32 2006