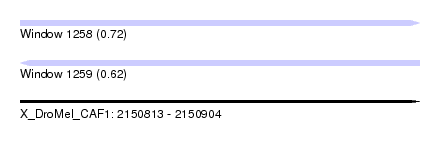
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,150,813 – 2,150,904 |
| Length | 91 |
| Max. P | 0.718290 |

| Location | 2,150,813 – 2,150,904 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
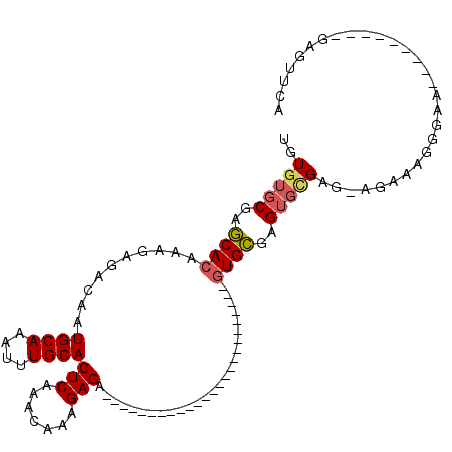
>X_DroMel_CAF1 2150813 91 + 22224390 UGUGUGCGAGCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGAGA-----------GCGAGUGCAA--GUGCGAGUGCGAG-AGAAAGGGAA---------GAGUUCA ..((..(..((((.....(......)....((((((((.............-----------..))))))))--))))..)..))..-..........---------....... ( -20.56) >DroSec_CAF1 30988 79 + 1 UGUGUGCGAGCACAAAGAGACGAUGCAAAUUUGCACUCAAACAAAGAGA-------------------------GUGCGAGUGCGAG-AGAAAGGGAA---------GAGUUCA (((((....))))).........((((...((((((((.........))-------------------------)))))).))))..-..........---------....... ( -18.90) >DroSim_CAF1 28489 79 + 1 UGUGUGCGAGCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGA-------------------------GUGCGAGUGCGAG-AGAAAGGGAA---------GAGUUCA (((((....))))).........((((...((((((((.........))-------------------------)))))).))))..-..........---------....... ( -18.90) >DroEre_CAF1 32104 79 + 1 UGUGUGCGAGCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGA-------------------------AUGCGAGUGCGAG-AGAAAGGGAA---------GAGUUCA (((((....)))))...............(((((((((...........-------------------------....)))))))))-..........---------....... ( -15.06) >DroYak_CAF1 34232 78 + 1 UGU-UGCGAGCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGA-------------------------GUGCGAGUGCGAG-AGAAAGGGAA---------GAGUUCA (.(-(((..(((...........)))..((((((((((.........))-------------------------)))))))))))).-).........---------....... ( -17.30) >DroAna_CAF1 37614 113 + 1 CGUG-GCAAUCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGACAAACAAGAUGAGACGAGCGAUGGUGUGGGAGGGUGAGGAGAUGGGGAAGUGGGGGAGCAGUUCA (.(.-((..(((((.........((((....))))(((.......))).........................)))))....)).).).......(((.((......)).))). ( -15.10) >consensus UGUGUGCGAGCACAAAGAGACAAUGCAAAUUUGCACUCAAACAAAGAGA_________________________GUGCGAGUGCGAG_AGAAAGGGAA_________GAGUUCA ..(((((..((((..........((((....))))(((.......)))..........................))))..)))))............................. (-13.10 = -13.38 + 0.28)

| Location | 2,150,813 – 2,150,904 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -14.95 |
| Consensus MFE | -8.14 |
| Energy contribution | -8.50 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
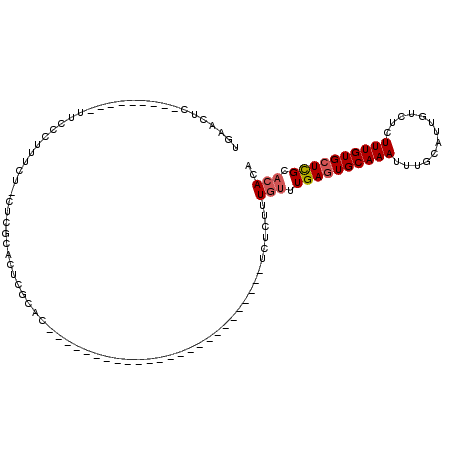
>X_DroMel_CAF1 2150813 91 - 22224390 UGAACUC---------UUCCCUUUCU-CUCGCACUCGCAC--UUGCACUCGC-----------UCUCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGCUCGCACACA .......---------..........-.........(((.--(((((((((.-----------.(.......)..)))))))))...)))..........((((....)))).. ( -18.70) >DroSec_CAF1 30988 79 - 1 UGAACUC---------UUCCCUUUCU-CUCGCACUCGCAC-------------------------UCUCUUUGUUUGAGUGCAAAUUUGCAUCGUCUCUUUGUGCUCGCACACA .......---------..........-...(((((((.((-------------------------.......)).)))))))..................((((....)))).. ( -15.60) >DroSim_CAF1 28489 79 - 1 UGAACUC---------UUCCCUUUCU-CUCGCACUCGCAC-------------------------UCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGCUCGCACACA .......---------..........-...(((((((.((-------------------------.......)).)))))))..................((((....)))).. ( -15.60) >DroEre_CAF1 32104 79 - 1 UGAACUC---------UUCCCUUUCU-CUCGCACUCGCAU-------------------------UCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGCUCGCACACA .......---------..........-...(((((((((.-------------------------......)))..))))))..................((((....)))).. ( -14.10) >DroYak_CAF1 34232 78 - 1 UGAACUC---------UUCCCUUUCU-CUCGCACUCGCAC-------------------------UCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGCUCGCA-ACA .......---------..........-...((....((((-------------------------((.........))))))......((((.........))))..)).-... ( -15.10) >DroAna_CAF1 37614 113 - 1 UGAACUGCUCCCCCACUUCCCCAUCUCCUCACCCUCCCACACCAUCGCUCGUCUCAUCUUGUUUGUCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGAUUGC-CACG ...........................................(((((......((.(..(((((((((.......))).))))))..)...)).......)))))...-.... ( -10.62) >consensus UGAACUC_________UUCCCUUUCU_CUCGCACUCGCAC_________________________UCUCUUUGUUUGAGUGCAAAUUUGCAUUGUCUCUUUGUGCUCGCACACA .......................................................................(((.((((((((((.............)))))))))).))).. ( -8.14 = -8.50 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:32 2006