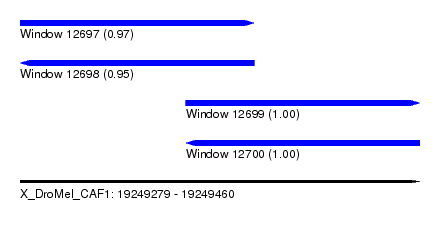
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,249,279 – 19,249,460 |
| Length | 181 |
| Max. P | 0.996702 |

| Location | 19,249,279 – 19,249,385 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -27.41 |
| Consensus MFE | -23.01 |
| Energy contribution | -22.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19249279 106 + 22224390 UUUCUAAGAGCGAUAUAUUUUGUCACUGCAUACUUUUUGACACUUGUUUUGCAAAUAAACGAUGAAAAUGGUCACAAAAGCCCAAGUGUCUAAAAGUAUGCAGCAA ...........((((.....)))).((((((((((((.((((((((....((......((.((....)).)).......)).)))))))).))))))))))))... ( -32.92) >DroSec_CAF1 36132 93 + 1 UUUC-------------UUUUGCCACUGCAUACUUUUUGACACUUGUUUUGCAAAUAAGCGAUGAAAAAGAUCACAAAAGCCCAAGUGCCCAAAAGUAUGCAGCAC ....-------------........((((((((((((.(.((((((..((((......))))(((......)))........)))))).).))))))))))))... ( -26.40) >DroSim_CAF1 37554 93 + 1 UUUC-------------UUUUGCCACUGCAUACUUUUUGACACUUGUUUUGCAAAUAAACGAUGAAAAAGAUCACAAAAGCCCAAGUGCCCAAAAGUAUGCAGCAA ....-------------........((((((((((((.(.(((((((((((...........(((......))))))))...)))))).).))))))))))))... ( -22.90) >consensus UUUC_____________UUUUGCCACUGCAUACUUUUUGACACUUGUUUUGCAAAUAAACGAUGAAAAAGAUCACAAAAGCCCAAGUGCCCAAAAGUAUGCAGCAA .........................((((((((((((.(.(((((((((.......))))(((.......)))..........))))).).))))))))))))... (-23.01 = -22.57 + -0.44)

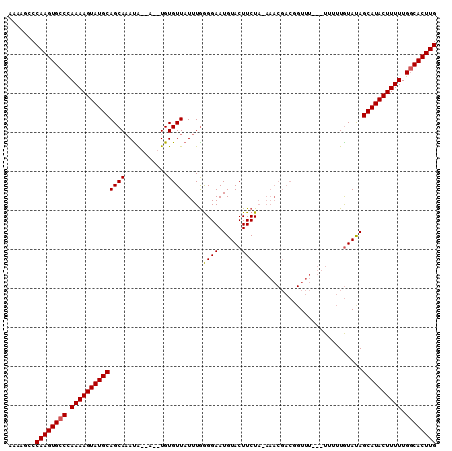
| Location | 19,249,279 – 19,249,385 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.40 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -5.38 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19249279 106 - 22224390 UUGCUGCAUACUUUUAGACACUUGGGCUUUUGUGACCAUUUUCAUCGUUUAUUUGCAAAACAAGUGUCAAAAAGUAUGCAGUGACAAAAUAUAUCGCUCUUAGAAA (..((((((((((((.(((((((((((......).)).........((......))....)))))))).))))))))))))..)...................... ( -34.20) >DroSec_CAF1 36132 93 - 1 GUGCUGCAUACUUUUGGGCACUUGGGCUUUUGUGAUCUUUUUCAUCGCUUAUUUGCAAAACAAGUGUCAAAAAGUAUGCAGUGGCAAAA-------------GAAA ..(((((((((((((.((((((((.((....(((((.......)))))......))....)))))))).))))))))))))).......-------------.... ( -36.10) >DroSim_CAF1 37554 93 - 1 UUGCUGCAUACUUUUGGGCACUUGGGCUUUUGUGAUCUUUUUCAUCGUUUAUUUGCAAAACAAGUGUCAAAAAGUAUGCAGUGGCAAAA-------------GAAA (..((((((((((((.((((((((...(((((..(.................)..))))))))))))).))))))))))))..).....-------------.... ( -34.33) >consensus UUGCUGCAUACUUUUGGGCACUUGGGCUUUUGUGAUCUUUUUCAUCGUUUAUUUGCAAAACAAGUGUCAAAAAGUAUGCAGUGGCAAAA_____________GAAA ..(((((((((((((.((((((((.......((((......)))).((......))....)))))))).)))))))))))))........................ (-33.84 = -33.40 + -0.44)

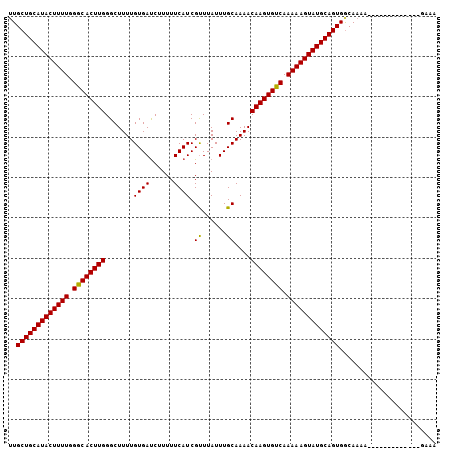
| Location | 19,249,354 – 19,249,460 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -25.29 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19249354 106 + 22224390 AAAAGCCCAAGUGUCUAAAAGUAUGCAGCAAAUA--A--UGUGUUAUUUAAGGAAUGUACUUCUAAAAACGACGGUUUCUUUAUUUGUACAGCAUACUUUUUGUCACUUG .......((((((.(.((((((((((.(((((((--(--..(((..(((..((((.....))))..)))..)))......))))))))...)))))))))).).)))))) ( -28.50) >DroSec_CAF1 36194 107 + 1 AAAAGCCCAAGUGCCCAAAAGUAUGCAGCACACAUUAUUUGUGUUAUUUGGGGAAUGUACUUCUAUAAACGACGGUUU---UUUUUGUAUAGCAUACUUUUUGGCACUUG .......((((((((.((((((((((((((((.......))))))..........(((((......((((....))))---.....))))))))))))))).)))))))) ( -35.90) >DroSim_CAF1 37616 92 + 1 AAAAGCCCAAGUGCCCAAAAGUAUGCAGCAAAUG--A--UGUGUUAUUUGGGGAAUGUACUUCU-----------UUU---UUUGUAUAUAGCAUACUUUUUGGCACUUG .......((((((((.((((((((((..((((((--(--....)))))))....((((((....-----------...---...)))))).)))))))))).)))))))) ( -34.30) >consensus AAAAGCCCAAGUGCCCAAAAGUAUGCAGCAAAUA__A__UGUGUUAUUUGGGGAAUGUACUUCUA_AAACGACGGUUU___UUUUUGUAUAGCAUACUUUUUGGCACUUG .......((((((((.((((((((((((((...........)))).....((((......))))...........................)))))))))).)))))))) (-25.29 = -25.40 + 0.11)

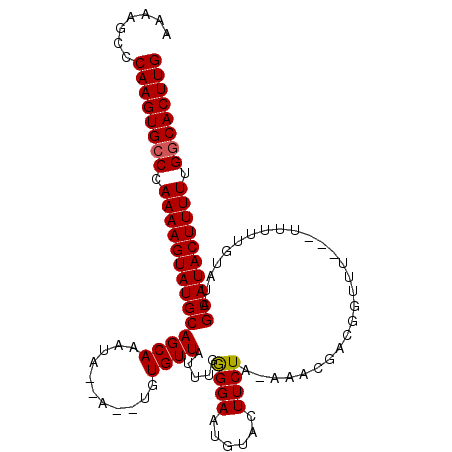
| Location | 19,249,354 – 19,249,460 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -23.84 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19249354 106 - 22224390 CAAGUGACAAAAAGUAUGCUGUACAAAUAAAGAAACCGUCGUUUUUAGAAGUACAUUCCUUAAAUAACACA--U--UAUUUGCUGCAUACUUUUAGACACUUGGGCUUUU ((((((.(.(((((((((((((((...((((((........))))))...))))).....(((((((....--)--))))))..)))))))))).).))))))....... ( -27.20) >DroSec_CAF1 36194 107 - 1 CAAGUGCCAAAAAGUAUGCUAUACAAAAA---AAACCGUCGUUUAUAGAAGUACAUUCCCCAAAUAACACAAAUAAUGUGUGCUGCAUACUUUUGGGCACUUGGGCUUUU ((((((((.((((((((((..........---((((....)))).....(((((((.....................))))))))))))))))).))))))))....... ( -32.90) >DroSim_CAF1 37616 92 - 1 CAAGUGCCAAAAAGUAUGCUAUAUACAAA---AAA-----------AGAAGUACAUUCCCCAAAUAACACA--U--CAUUUGCUGCAUACUUUUGGGCACUUGGGCUUUU ((((((((.((((((((((..........---...-----------.(((.....)))..(((((......--.--.)))))..)))))))))).))))))))....... ( -28.00) >consensus CAAGUGCCAAAAAGUAUGCUAUACAAAAA___AAACCGUCGUUU_UAGAAGUACAUUCCCCAAAUAACACA__U__UAUUUGCUGCAUACUUUUGGGCACUUGGGCUUUU ((((((((.((((((((((............................(((.....)))..((((((..........))))))..)))))))))).))))))))....... (-23.84 = -24.40 + 0.56)

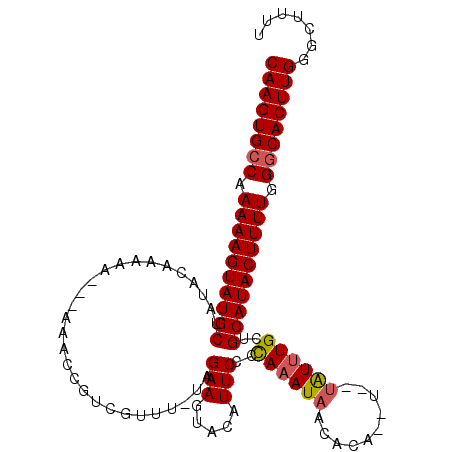
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:24 2006