
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,248,801 – 19,248,966 |
| Length | 165 |
| Max. P | 0.847788 |

| Location | 19,248,801 – 19,248,906 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -11.85 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248801 105 + 22224390 CCCACUUGCCACCACCCACUUGCC-ACCA-CCCACUGCUGUUUGCUGUCUCAGCGCAAAAUGUUGCUGCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAU .....................((.-....-......((((..........))))((((....)))).))......((((((((((.........))))))))))... ( -15.70) >DroPse_CAF1 79100 93 + 1 --------------CCUGCCUGCCUGCCGGCCUUUCAGUGUUUGCUGGCUCAGCGCAAAACGCUGCUGCUCUAAUUAUAAUGAAACGAUUUUCAUUUCAUUUUGCAU --------------..((((((...((((((............)))))).))).))).........(((...(((...(((((((....)))))))..)))..))). ( -21.30) >DroSec_CAF1 35670 98 + 1 CCC-------ACCACCCACUUGCC-ACCA-CCCACUGCUGUUUGCUGUCUCAGCGCAAAAUGUUGCUGCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAU ...-------...........((.-....-......((((..........))))((((....)))).))......((((((((((.........))))))))))... ( -15.70) >DroEre_CAF1 33081 96 + 1 GGC-------ACCACCCACUUG----CCACCCCACUGCUGUUUGCUGUCUCAGCGCAAAAUGUUGCUGCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAU (((-------(.........))----))........((.....((((...))))((((....)))).))......((((((((((.........))))))))))... ( -19.70) >DroYak_CAF1 34921 97 + 1 --C-------ACCACCCACUAGCU-ACCACCCCACUGCUGUUUGCUGUCUCAGCGCAAAAUGUUGCUGCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAU --.-------..........(((.-...........((((..........))))((((....)))).))).....((((((((((.........))))))))))... ( -16.80) >DroPer_CAF1 46192 93 + 1 --------------CCUGCCUGCCUGCCGGCCUUUCAGUGUUUGCUGGCUCAGCGCAAAACGCUGCUGCUCUAAUUAUAAUGAAACGAUUUUCAUUUCAUUUUGCAU --------------..((((((...((((((............)))))).))).))).........(((...(((...(((((((....)))))))..)))..))). ( -21.30) >consensus __C_______ACCACCCACUUGCC_ACCA_CCCACUGCUGUUUGCUGUCUCAGCGCAAAAUGUUGCUGCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAU ...........................................((((...))))(((......))).........((((((((((.........))))))))))... (-11.85 = -11.97 + 0.11)

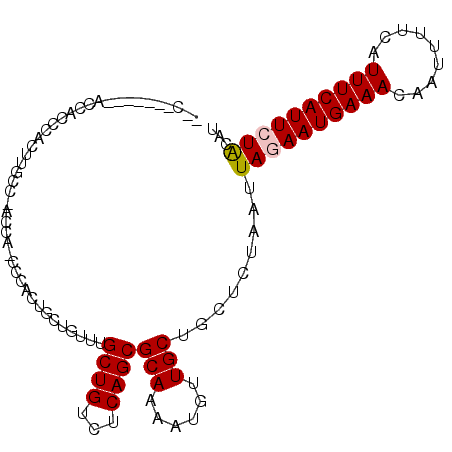
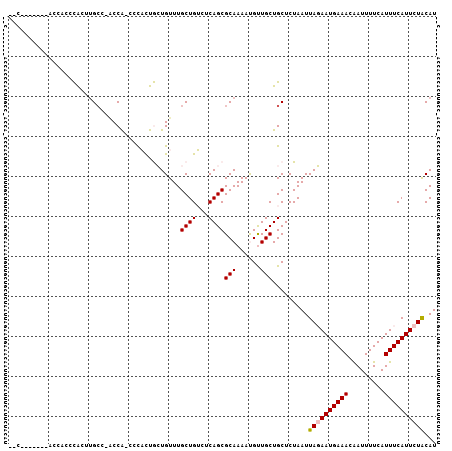
| Location | 19,248,801 – 19,248,906 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.34 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -17.19 |
| Energy contribution | -17.58 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248801 105 - 22224390 AUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGCAGUGGG-UGGU-GGCAAGUGGGUGGUGGCAAGUGGG ...((((((((((..(...)..))))))))))......((.((.((..(((((((((...))))...((.((.....-)).)-))))))...)).)).))....... ( -27.10) >DroPse_CAF1 79100 93 - 1 AUGCAAAAUGAAAUGAAAAUCGUUUCAUUAUAAUUAGAGCAGCAGCGUUUUGCGCUGAGCCAGCAAACACUGAAAGGCCGGCAGGCAGGCAGG-------------- .(((..(((((((((.....))))))))).......(.((..(((((.....))))).))).)))....(((....(((....)))...))).-------------- ( -29.60) >DroSec_CAF1 35670 98 - 1 AUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGCAGUGGG-UGGU-GGCAAGUGGGUGGU-------GGG ...((((((((((..(...)..)))))))))).......((.((.(((((.((((((...))))...((.((.....-)).)-))))))))..)).)-------).. ( -25.70) >DroEre_CAF1 33081 96 - 1 AUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGCAGUGGGGUGG----CAAGUGGGUGGU-------GCC ...((((((((((..(...)..))))))))))......(((.((.(((((.((((((...))))...((.(.....).)))----))))))..)).)-------)). ( -27.70) >DroYak_CAF1 34921 97 - 1 AUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGCAGUGGGGUGGU-AGCUAGUGGGUGGU-------G-- ...((((((((((..(...)..))))))))))((((..(((((..(((((..(((((..........)))).)..)))))..-.))).))...))))-------.-- ( -26.00) >DroPer_CAF1 46192 93 - 1 AUGCAAAAUGAAAUGAAAAUCGUUUCAUUAUAAUUAGAGCAGCAGCGUUUUGCGCUGAGCCAGCAAACACUGAAAGGCCGGCAGGCAGGCAGG-------------- .(((..(((((((((.....))))))))).......(.((..(((((.....))))).))).)))....(((....(((....)))...))).-------------- ( -29.60) >consensus AUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGCAGUGGG_UGGU_GGCAAGUGGGUGGU_______G__ ...(((((((((((((...)))))))))))))......((.((((....))))((((...))))....................))..................... (-17.19 = -17.58 + 0.39)


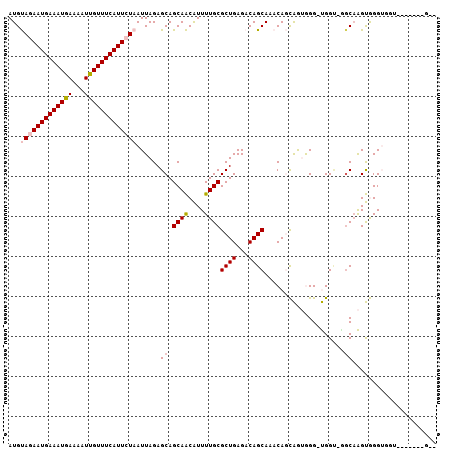
| Location | 19,248,836 – 19,248,935 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -13.64 |
| Energy contribution | -14.17 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248836 99 - 22224390 -----------AAGGCAAUGCCAACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAG---------- -----------..(((...)))..........((((....))))(((((((((..(...)..)))))))))..........((((....))))((((...))))......---------- ( -22.40) >DroVir_CAF1 73416 102 - 1 -----------GAGGCAAAAGCAA--ACGACAACAUGAAAAUGCAAA-----AUGAAAAUCGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAGACGUCAAACA -----------...((....))..--..(((...........(((((-----(((......(((..(.......)..))).....))))))))((((...))))........)))..... ( -17.20) >DroPse_CAF1 79123 99 - 1 -----------ACGGCUAUGGAAGCGACGACAACAUGAAAAUGCAAAAUGAAAUGAAAAUCGUUUCAUUAUAAUUAGAGCAGCAGCGUUUUGCGCUGAGCCAGCAAACAC---------- -----------..((((......((........(((....)))...(((((((((.....))))))))).........))..(((((.....))))))))).........---------- ( -26.80) >DroEre_CAF1 33107 110 - 1 GUGGAAGCAGCAUGGCAAUGCCAACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAG---------- ......((((.(((....((((..........((((....))))(((((((((..(...)..))))))))).....).)))....))).))))((((...))))......---------- ( -27.60) >DroYak_CAF1 34948 95 - 1 ---------------CAACGACGACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAG---------- ---------------.................((((....))))(((((((((..(...)..)))))))))..........((((....))))((((...))))......---------- ( -19.50) >DroPer_CAF1 46215 99 - 1 -----------ACGGCUAUGGAAGCGACGACAACAUGAAAAUGCAAAAUGAAAUGAAAAUCGUUUCAUUAUAAUUAGAGCAGCAGCGUUUUGCGCUGAGCCAGCAAACAC---------- -----------..((((......((........(((....)))...(((((((((.....))))))))).........))..(((((.....))))))))).........---------- ( -26.80) >consensus ___________AAGGCAAUGGCAACGACGACAACAUGAAAAUGCAAAAUGAAAUGAAAAUCGUUUCAUUCUAAUUAGAGCAGCAACAUUUUGCGCUGAGACAGCAAACAG__________ ..............((.................(((....)))...((((((((((...)))))))))).........)).((((....))))((((...))))................ (-13.64 = -14.17 + 0.53)
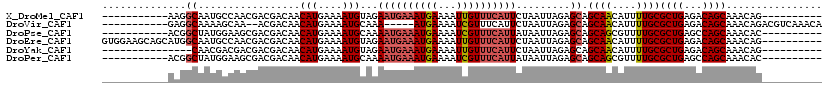
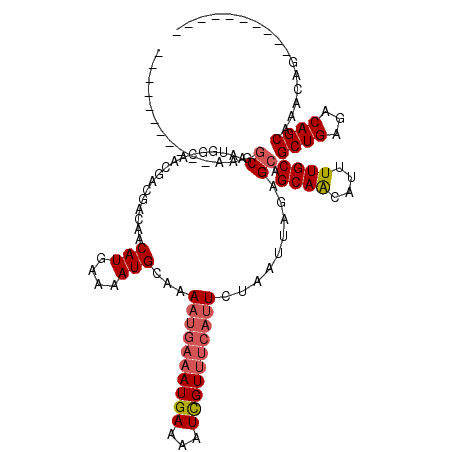
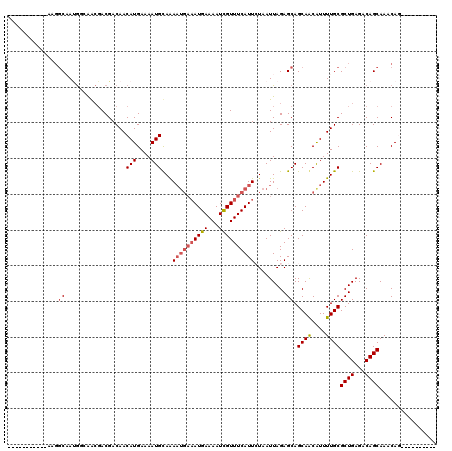
| Location | 19,248,866 – 19,248,966 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -14.21 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248866 100 + 22224390 GCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAUUUUCAUGUUGUCGUCGUUGGCAUUGCCUU-------------ACUGCU------UGCUACACGGAUAAAAA-GCAGCUCU (((((...((((((((((.........))))))))))..........(((((..((((((((..((...-------------...)).------))))).))))))))..)-).)))... ( -21.80) >DroSec_CAF1 35728 106 + 1 GCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAUUUUCAUGUUAUCGUCGUUGGCAUUGCCUU-------------ACUGCUCGCUUUUGCUACACGGAUAAAAA-GCAGCUCU (((((...((((((((((.........))))))))))..........(((((..((((((((..((...-------------.......))...))))).))))))))..)-).)))... ( -21.20) >DroEre_CAF1 33137 113 + 1 GCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAUUUUCAUGUUGUCGUCGUUGGCAUUGCCAUGCUGCUUCCACUUACUGCU------UGCUACACGGAUAAAAA-GCAGCUCU ((......((((((((((.........))))))))))...........(((((....)))))..))...(((((((((..........------........))).....)-)))))... ( -21.47) >DroYak_CAF1 34978 97 + 1 GCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAUUUUCAUGUUGUCGUCGUCGUCGUUG-----------------GCUGCU------UGCUACACGGAUAAAAAGGCAGCUCU ........((((((((((.........)))))))))).........((((((.......((((((-----------------((....------.)))).)))).......))))))... ( -23.14) >consensus GCUCUAAUUAGAAUGAAACAAUUUUCAUUUCAUUCUACAUUUUCAUGUUGUCGUCGUUGGCAUUGCCUU_____________ACUGCU______UGCUACACGGAUAAAAA_GCAGCUCU (((.....((((((((((.........))))))))))..........(((((..(((.(((...)))..................((........))...))))))))......)))... (-14.21 = -14.53 + 0.31)

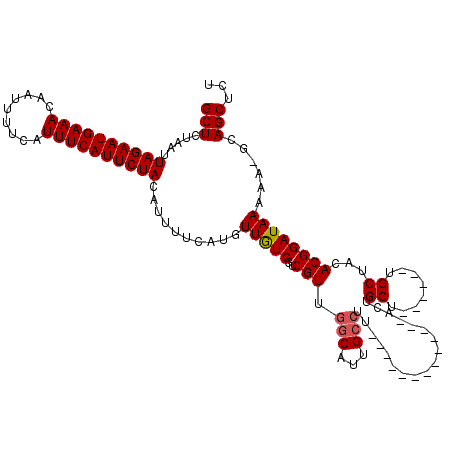
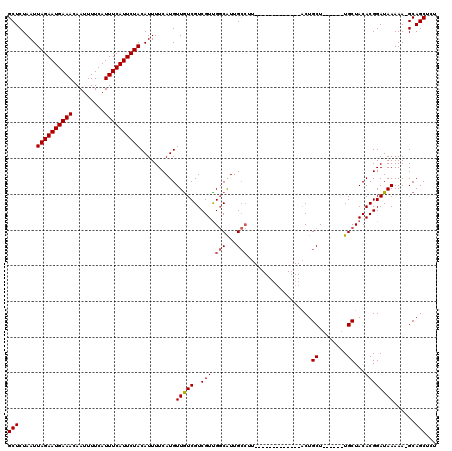
| Location | 19,248,866 – 19,248,966 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248866 100 - 22224390 AGAGCUGC-UUUUUAUCCGUGUAGCA------AGCAGU-------------AAGGCAAUGCCAACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGC ...(((((-((.((((....)))).)------))))))-------------..(((...)))..........((((....))))(((((((((..(...)..)))))))))......... ( -25.20) >DroSec_CAF1 35728 106 - 1 AGAGCUGC-UUUUUAUCCGUGUAGCAAAAGCGAGCAGU-------------AAGGCAAUGCCAACGACGAUAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGC ...(((((-((..(((....)))((....)))))))))-------------..(((...)))..........((((....))))(((((((((..(...)..)))))))))......... ( -23.70) >DroEre_CAF1 33137 113 - 1 AGAGCUGC-UUUUUAUCCGUGUAGCA------AGCAGUAAGUGGAAGCAGCAUGGCAAUGCCAACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGC ...(((((-((((......(((....------.)))......))))))))).((((...)))).........((((....))))(((((((((..(...)..)))))))))......... ( -30.20) >DroYak_CAF1 34978 97 - 1 AGAGCUGCCUUUUUAUCCGUGUAGCA------AGCAGC-----------------CAACGACGACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGC .(.(((((.((.((((....)))).)------))))))-----------------)................((((....))))(((((((((..(...)..)))))))))......... ( -20.30) >consensus AGAGCUGC_UUUUUAUCCGUGUAGCA______AGCAGU_____________AAGGCAAUGCCAACGACGACAACAUGAAAAUGUAGAAUGAAAUGAAAAUUGUUUCAUUCUAAUUAGAGC ...(((...........(((((.((........))..................(((...)))..........)))))......((((((((((..(...)..)))))))))).....))) (-18.27 = -18.77 + 0.50)

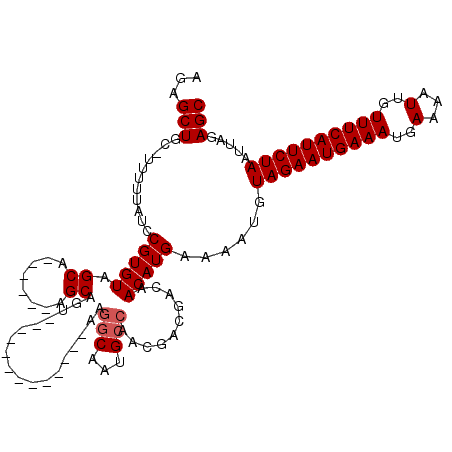
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:21 2006