| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,248,461 – 19,248,605 |
| Length | 144 |
| Max. P | 0.998429 |

| Location | 19,248,461 – 19,248,569 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
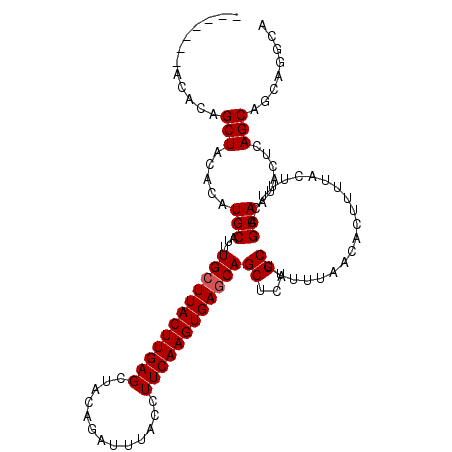
>X_DroMel_CAF1 19248461 108 + 22224390 UGAGCACAUAAGUGUCAUAAUCACACACACACAUAGCUACACAUAGCUACACAUGCAUUUGCUUACUUGAGCUAGAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUU (((((.(((..((((.......)))).......((((((....))))))...)))....((((((((((((.............)))))))))))))))))....... ( -29.32) >DroSec_CAF1 35335 94 + 1 UGAGCACAUAAGUGUCAUAAUCACACACAC--------------AGCUACACAUGCAUUUGCUUACUUGAGCUAGAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUU (((((......((((.......))))....--------------.((.......))...((((((((((((.............)))))))))))))))))....... ( -25.32) >DroEre_CAF1 32737 94 + 1 UGAGCACAUAAGUGUCAUAAUCACAC--------------ACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAACAGCUCAGCUUUUU (((((......((((.......))))--------------.....((.......))...((.(((((((((.............))))))))).)))))))....... ( -18.72) >DroYak_CAF1 34574 102 + 1 UGAGCACAUAAGUGUCAUAAUCACACACACACA------CACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUU (((((......((((.......)))).......------......((.......))...((((((((((((.............)))))))))))))))))....... ( -25.32) >consensus UGAGCACAUAAGUGUCAUAAUCACACACAC__________ACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUU (((((......((((.......))))...................((.......))...((((((((((((.............)))))))))))))))))....... (-23.27 = -23.52 + 0.25)


| Location | 19,248,494 – 19,248,605 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -17.30 |
| Energy contribution | -17.55 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19248494 111 + 22224390 UAGCUACACAUAGCUACACAUGCAUUUGCUUACUUGAGCUAGAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUUAACACUUUUACUUUCAGCAAUACUCAGCAGCGGUGG ((((((....))))))(((.(((..(((((((((((((.............)))))))))))))....(((...................)))........)))...))). ( -27.13) >DroSec_CAF1 35365 100 + 1 -----------AGCUACACAUGCAUUUGCUUACUUGAGCUAGAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUUCACACUUUUACUUUCAGCAAUACUCAGCAGCGGUGG -----------..((((...(((..(((((((((((((.............)))))))))))))....(((...................)))........)))...)))) ( -22.53) >DroEre_CAF1 32763 104 + 1 -------ACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAACAGCUCAGCUUUUUAACACUUUUACUUUCAGCAAUACUCAGCAGUAGGCA -------.....((.......))...((((((((((((((...(.(((((......)))))))))))).......................((........)))))))))) ( -19.70) >DroYak_CAF1 34607 105 + 1 ------CACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUUAACACUUUUACUUUCAGCAAUACUCAGCAGUAGGCA ------......(((..((.(((..(((((((((((((.............)))))))))))))....(((...................)))........))))).))). ( -21.73) >consensus _______ACACAGCUACACAUGCAUUUGCUUACUUGAGCUACAGAUUUACCUUCAAGUGAGCAGCUCAGCUUUUUAACACUUUUACUUUCAGCAAUACUCAGCAGCAGGCA ............(((.....(((...((((((((((((.............))))))))))))((...)).....................)))......)))........ (-17.30 = -17.55 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:17 2006