
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,243,142 – 19,243,322 |
| Length | 180 |
| Max. P | 0.896502 |

| Location | 19,243,142 – 19,243,247 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.39 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -18.59 |
| Energy contribution | -19.21 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649539 |
| Prediction | RNA |
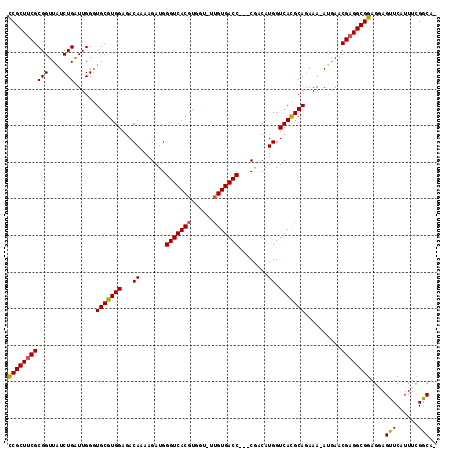
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19243142 105 - 22224390 UGACCAUGUCGGGUCACAAAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGCUGCCACUACCACUACCCCCAACCUCUCG .(((...(..(((((((........)))))))..)....)))..............(((....((((...))))..))).......................... ( -21.40) >DroSec_CAF1 30078 98 - 1 UGACCAUGUCGGGUCACAA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGCAGUCACUCCC----CCCCCAC--CCUCG ((((..(((((((((((..-.....)))))))......(((......)))........)))).((((...))))....)))).....----.......--..... ( -23.40) >DroSim_CAF1 26657 97 - 1 UGACCAUGUCGGGUCACCA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGCAGUCACUCCC-----CCCCAC--CCUCG ((((..(((((((((((..-.....)))))))......(((......)))........)))).((((...))))....)))).....-----......--..... ( -22.80) >DroEre_CAF1 27888 89 - 1 UGACCAUGCUGGGUCACAA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAACCAGACAACCGCGAAGCGAAGCAGCCACUCCC----CUU----------- ......(((((((((((..-.....))))))).....((((((..............)))))).(((...))).)))).........----...----------- ( -21.14) >consensus UGACCAUGUCGGGUCACAA_ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGGAGCAGCCACUCCC____CCCCCAC__CCUCG ((((..(((.(((((((........))))))).....((((((..............))))))((((...)))).)))))))....................... (-18.59 = -19.21 + 0.62)


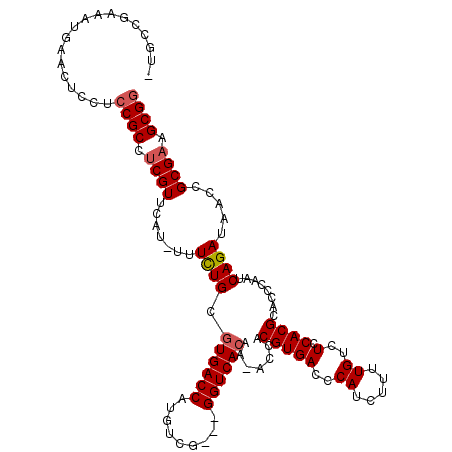
| Location | 19,243,173 – 19,243,286 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -38.91 |
| Consensus MFE | -33.46 |
| Energy contribution | -33.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19243173 113 + 22224390 CCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGUUUUGUGACC---CGACAUGGUCAUGCAGAAA-AGGAACGAGGCGGAGUAGUUCAUUUCGGCAG (((((((((((((....)))))..(((((((...((...(.(((((((((......)))))))---)).).)).)))))))....-.....)))))))).................. ( -37.80) >DroSec_CAF1 30103 111 + 1 CCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGU-UUGUGACC---CGACAUGGUCACGCAGAAA-AUAAACGAGGCGGAGGAGUUCAUUUCGGCA- (((((((((((((....)))))..(((((((...((...(.(((((((((....-.)))))))---)).).)).)))))))....-.....))))))))..(((.....)))....- ( -39.60) >DroSim_CAF1 26681 111 + 1 CCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGU-UGGUGACC---CGACAUGGUCACGCAGAAA-AUAAACGAGGCGGAGGAGUUCAUUUUGGCG- (((((((((((((....)))))..(((((((...((...(.((((((((.....-..))))))---)).).)).)))))))....-.....)))))))).................- ( -38.20) >DroEre_CAF1 27904 112 + 1 UCGCUUCGCGGUUGUCUGGUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGU-UUGUGACC---CAGCAUGGUCACGCAAAAAGAUGAACGAGGCGGAGAAGCUCAUUUCGGCA- ((((((((...(..(((..(((.((((.((....))...(.(((((((((....-.)))))))---)).)...).))).)))..)))..).))))))))....(((......))).- ( -44.80) >DroYak_CAF1 29319 115 + 1 CCGCUACGCGGUUAUCUGGUUGGAUGCGUGGAGACAAAAGAUGGGUCACGUGGU-UUGUGACCAGACACCAUGGUCACGCAGAAAAAUGAACGAGGCGGAGGAGUUCAUUUCGGCG- ((((...)))).......((((..(((((((...((.......(((((((....-.)))))))........)).)))))))...((((((((...........)))))))))))).- ( -34.16) >consensus CCGCUUCGCGGUUAUCUGAUUGGGUGCGUGGAGACAAAAGAUGGGUCACGUGGU_UUGUGACC___CGACAUGGUCACGCAGAAA_AUGAACGAGGCGGAGGAGUUCAUUUCGGCA_ (((((((((((....)))......(((((((...((.......(((((((......)))))))........)).)))))))..........))))))))....(((......))).. (-33.46 = -33.38 + -0.08)

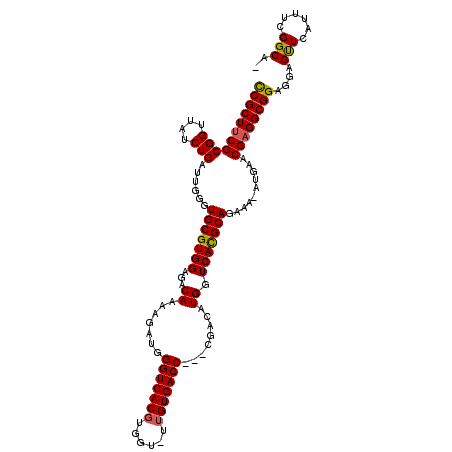
| Location | 19,243,173 – 19,243,286 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.77 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19243173 113 - 22224390 CUGCCGAAAUGAACUACUCCGCCUCGUUCCU-UUUCUGCAUGACCAUGUCG---GGUCACAAAACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGG ..................((((.((((....-..((((...(((...)))(---((((((........)))))))........................)))).....)))).)))) ( -28.10) >DroSec_CAF1 30103 111 - 1 -UGCCGAAAUGAACUCCUCCGCCUCGUUUAU-UUUCUGCGUGACCAUGUCG---GGUCACAA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGG -.................((((.((((((((-((..((((((..((.(..(---((((((..-.....)))))))..)...))....)))))).......))))))..)))).)))) ( -30.90) >DroSim_CAF1 26681 111 - 1 -CGCCAAAAUGAACUCCUCCGCCUCGUUUAU-UUUCUGCGUGACCAUGUCG---GGUCACCA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGG -.................((((.((((((((-((..((((((..((.(..(---((((((..-.....)))))))..)...))....)))))).......))))))..)))).)))) ( -30.30) >DroEre_CAF1 27904 112 - 1 -UGCCGAAAUGAGCUUCUCCGCCUCGUUCAUCUUUUUGCGUGACCAUGCUG---GGUCACAA-ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAACCAGACAACCGCGAAGCGA -...........(((((..((....(((........((((((..((.(.((---((((((..-.....)))))))).)...))....))))))........)))...)).))))).. ( -28.99) >DroYak_CAF1 29319 115 - 1 -CGCCGAAAUGAACUCCUCCGCCUCGUUCAUUUUUCUGCGUGACCAUGGUGUCUGGUCACAA-ACCACGUGACCCAUCUUUUGUCUCCACGCAUCCAACCAGAUAACCGCGUAGCGG -....(((((((((...........)))))))))..((((((..((.((((...((((((..-.....))))))))))...))....)))))).............((((...)))) ( -29.50) >consensus _UGCCGAAAUGAACUCCUCCGCCUCGUUCAU_UUUCUGCGUGACCAUGUCG___GGUCACAA_ACCACGUGACCCAUCUUUUGUCUCCACGCACCCAAUCAGAUAACCGCGAAGCGG ..................((((.((((.......((((.((((((.........)))))).......(((((..((.....))..).))))........)))).....)))).)))) (-22.18 = -22.62 + 0.44)


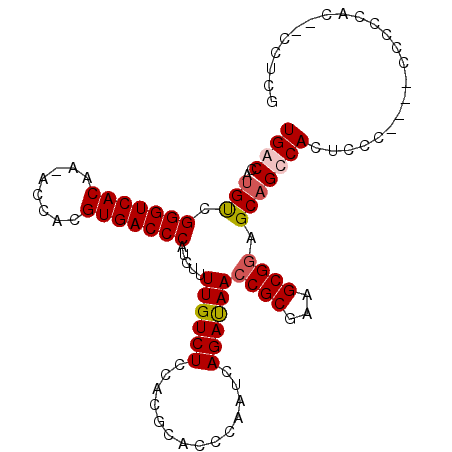
| Location | 19,243,210 – 19,243,322 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -30.44 |
| Consensus MFE | -19.72 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19243210 112 - 22224390 CUUGUACG----CUUACAGGGUAUUUAAGUCUCUGCCUGCCUGCCGAAAUGAACUACUCCGCCUCGUUCCU-UUUCUGCAUGACCAUGUCG---GGUCACAAAACCACGUGACCCAUCUU .......(----(...(((((.........)))))...)).(((.((((.((((...........))))..-)))).))).(((...)))(---((((((........)))))))..... ( -27.80) >DroSec_CAF1 30140 107 - 1 UUUGUACG----CUUACAGGGUAUUUAAAUCUCCGCG----UGCCGAAAUGAACUCCUCCGCCUCGUUUAU-UUUCUGCGUGACCAUGUCG---GGUCACAA-ACCACGUGACCCAUCUU ..((((..----..))))((((...........((((----((..(((((((((...........))))))-)))....((((((......---))))))..-..))))))))))..... ( -27.10) >DroSim_CAF1 26718 100 - 1 -------G----GUUACAGGGUAUUUAAAUCUCUGCC----CGCCAAAAUGAACUCCUCCGCCUCGUUUAU-UUUCUGCGUGACCAUGUCG---GGUCACCA-ACCACGUGACCCAUCUU -------(----(((((.(((((..........))))----)((.(((((((((...........))))))-)))..)))))))).....(---((((((..-.....)))))))..... ( -33.80) >DroEre_CAF1 27941 107 - 1 -UUGUACG----CUUACAGGGUAUUCAAACCUCUGCC----UGCCGAAAUGAGCUUCUCCGCCUCGUUCAUCUUUUUGCGUGACCAUGCUG---GGUCACAA-ACCACGUGACCCAUCUU -...((((----(...((((((......))).)))..----....(((.(((((......).)))))))........))))).......((---((((((..-.....)))))))).... ( -29.10) >DroYak_CAF1 29356 114 - 1 -UUGCAAGAUGUUUUACAGGGUAUUUAAAUCUCUGCC----CGCCGAAAUGAACUCCUCCGCCUCGUUCAUUUUUCUGCGUGACCAUGGUGUCUGGUCACAA-ACCACGUGACCCAUCUU -....((((((..((((.(((((..........))))----)((.(((((((((...........)))))))))...))(((((((.......)))))))..-.....))))..)))))) ( -34.40) >consensus _UUGUACG____CUUACAGGGUAUUUAAAUCUCUGCC____UGCCGAAAUGAACUCCUCCGCCUCGUUCAU_UUUCUGCGUGACCAUGUCG___GGUCACAA_ACCACGUGACCCAUCUU ..................((((....................((.(((((((((...........)))))..)))).))((((((.........))))))...........))))..... (-19.72 = -19.80 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:14 2006