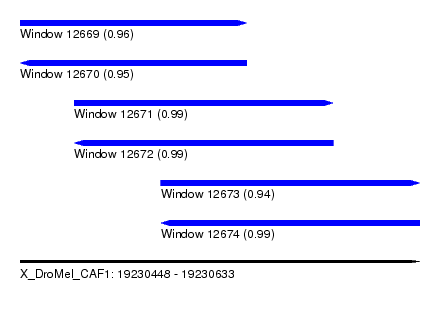
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,230,448 – 19,230,633 |
| Length | 185 |
| Max. P | 0.994423 |

| Location | 19,230,448 – 19,230,553 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 96.21 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -41.22 |
| Energy contribution | -41.55 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230448 105 + 22224390 UGGUUUGGUUUAGUAU-GGGUAUGGUGGCUAGCCUGGGCCAUGGGGCAAUACGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUC ......(((((.((((-..((.(.((((((......)))))).).)).)))).)))))((((((.....))))))((.((((((((........)))))))).)). ( -38.30) >DroSec_CAF1 17491 104 + 1 UGGUUUGGUUUAGUA--UGGUAUGGCGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUC (((((..(((((((.--..(.....).)))))))..)))))((((..((((.........))))..)))).....((.((((((((........)))))))).)). ( -43.80) >DroSim_CAF1 17159 106 + 1 UGGUUUGGUUUAGUAUUUGGUAUGGUGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUC (((((..((((((((((......))).)))))))..)))))((((..((((.........))))..)))).....((.((((((((........)))))))).)). ( -44.00) >consensus UGGUUUGGUUUAGUAU_UGGUAUGGUGGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUC (((((..(((((((.............)))))))..)))))..((((.........((((((((.....)))))))).((((((((........)))))))))))) (-41.22 = -41.55 + 0.33)


| Location | 19,230,448 – 19,230,553 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 96.21 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230448 105 - 22224390 GAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGUAUUGCCCCAUGGCCCAGGCUAGCCACCAUACCC-AUACUAAACCAAACCA .(((((((((((........)))))))(((......((((.(((((........).)))).)))).)))...)))).............-................ ( -30.60) >DroSec_CAF1 17491 104 - 1 GAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCGCCAUACCA--UACUAAACCAAACCA (((.((((((((........)))))))).))).(((((((..........(....)............)))))))..............--............... ( -32.35) >DroSim_CAF1 17159 106 - 1 GAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCACCAUACCAAAUACUAAACCAAACCA (((.((((((((........)))))))).))).(((((((..........(....)............)))))))............................... ( -32.35) >consensus GAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCCACCAUACCA_AUACUAAACCAAACCA (((.((((((((........)))))))).))).(((((((.........((........)).......)))))))............................... (-30.32 = -30.66 + 0.33)



| Location | 19,230,473 – 19,230,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -48.50 |
| Consensus MFE | -45.50 |
| Energy contribution | -45.18 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230473 120 + 22224390 GGCUAGCCUGGGCCAUGGGGCAAUACGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCC ((((.((..(((...((((((.........((((((((.....)))))))).((((((((........))))))))))))))))).)).))))((((..((.(......)))..)))).. ( -48.20) >DroSec_CAF1 17515 120 + 1 GGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAGGUGGGCGGUGUUACUUUGCUUUCC (((..(((((((...(..(((........)))..).....)))))))((((.((((((((........))))))))((.((((((((.....)))))))))).......))))))).... ( -51.50) >DroSim_CAF1 17185 120 + 1 GGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCC ((((.(...(((...((((((.........((((((((.....)))))))).((((((((........)))))))))))))))))..).))))((((..((.(......)))..)))).. ( -45.70) >DroEre_CAF1 15690 120 + 1 GGCUAGACGGGGCUAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUUCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCC ((((.(.(((((...((((((.........((((((((.....)))))))).((((((((........)))))))))))))))))))).))))((((..((.(......)))..)))).. ( -45.50) >DroYak_CAF1 16419 120 + 1 GGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUAGCAAGCCAAAGUGGGCGGUGUUACUUCGCUUUCC ((((.(.(((((...((((((.........((((((((.....)))))))).((((((((........)))))))))))))))))))).))))...(..(((((.......)))))..). ( -51.60) >consensus GGCUAGACUGGGCCAUGGGGCAAUCCGAAGCUUUGAUUAUCCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCC ((((.(.(.(((...((((((.........((((((((.....)))))))).((((((((........))))))))))))))))).)).))))...(..(((((.......)))))..). (-45.50 = -45.18 + -0.32)


| Location | 19,230,473 – 19,230,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -44.98 |
| Consensus MFE | -42.43 |
| Energy contribution | -42.91 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230473 120 - 22224390 GGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGUAUUGCCCCAUGGCCCAGGCUAGCC ((..(((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))..(((((.........((........)).......))))))))..)) ( -45.29) >DroSec_CAF1 17515 120 - 1 GGAAAGCAAAGUAACACCGCCCACCUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCC .....((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))(((((((..........(....)............)))))))..)). ( -48.65) >DroSim_CAF1 17185 120 - 1 GGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCC .....((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))(((((((..........(....)............)))))))..)). ( -45.95) >DroEre_CAF1 15690 120 - 1 GGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGAAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUAGCCCCGUCUAGCC ((...((..(((......((((((((((.....)))))))).))(((((((((............))))))))))))(((.(((((.........))))).)))...)).))........ ( -38.80) >DroYak_CAF1 16419 120 - 1 GGAAAGCGAAGUAACACCGCCCACUUUGGCUUGCUAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCC ...(((((((((..........))))).))))(((((((((.((((((((((........))))))).........((((.(((((.........))))).)))).)))))).)))))). ( -46.20) >consensus GGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGGAUAAUCAAAGCUUCGGAUUGCCCCAUGGCCCAGUCUAGCC .....((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))(((((((.........((........)).......)))))))..)). (-42.43 = -42.91 + 0.48)

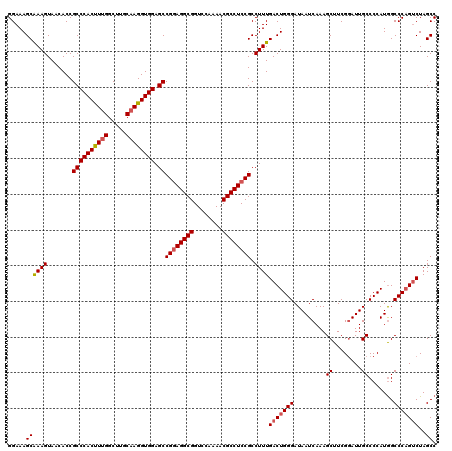
| Location | 19,230,513 – 19,230,633 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -36.73 |
| Energy contribution | -36.61 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230513 120 + 22224390 CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAUACCCAGGACCAUUUGUUUUUGGCUCCUUUGUGAUUU ...((.(((((.((((((((........))))))))((.((((.(((.....))).)))))).......))))))).....((((...(((((((........))).)))).)))).... ( -39.10) >DroSec_CAF1 17555 120 + 1 CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAGGUGGGCGGUGUUACUUUGCUUUCCCCAUCCCCAGCACCAUUUGUUUUCGGCUCCUUUGUGAUUU ......((((((((((((((........))))))))((.((((((((.....))))))))))((((((....((.......)).....))))))..............))))))...... ( -43.40) >DroSim_CAF1 17225 120 + 1 CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAUCCCCAGCACCAUUUGUUUUCGGCUCCUUUGUGAUUU ......((((((((((((((........))))))))((........)).((((((((..((.((((((....((.......)).....)))))).))..)))).))))))))))...... ( -38.50) >DroEre_CAF1 15730 114 + 1 CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUUCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCC------CCAGCACCAUUUGUUUUCGGCUCCUUUGUGAUUC ......((((((((((((((........))))))))((........)).((((((((..((.((((((............------..)))))).))..)))).))))))))))...... ( -35.74) >DroYak_CAF1 16459 114 + 1 CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUAGCAAGCCAAAGUGGGCGGUGUUACUUCGCUUUCC------CCAGCACCAUUUGUUUUCGGCUCCUUUGUGAUUU ......((((((((((((((........))))))))(((......))).((((((((..((.((((((............------..)))))).))..)))).))))))))))...... ( -38.94) >consensus CCCAGUCAAAGGCGGAGGCGUUUUGGACCGCCUCCGGCUCCACCUUGCAAGCCAAAGUGGGCGGUGUUACUUUGCUUUCCCCAU_CCCAGCACCAUUUGUUUUCGGCUCCUUUGUGAUUU ......((((((((((((((........))))))))((........)).((((((((..((.((((((....................)))))).))..)))).))))))))))...... (-36.73 = -36.61 + -0.12)

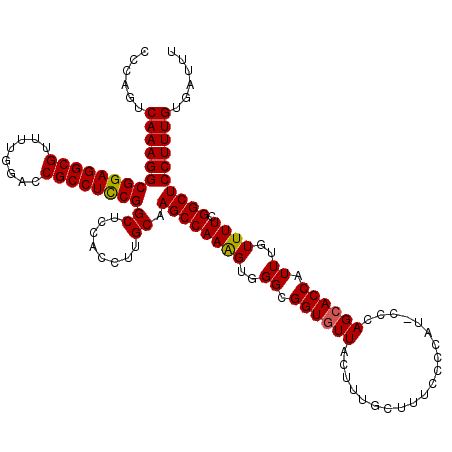
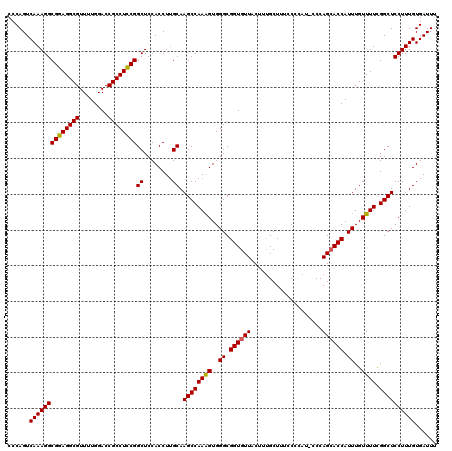
| Location | 19,230,513 – 19,230,633 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.47 |
| Mean single sequence MFE | -40.54 |
| Consensus MFE | -38.08 |
| Energy contribution | -38.36 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19230513 120 - 22224390 AAAUCACAAAGGAGCCAAAAACAAAUGGUCCUGGGUAUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG .....((..((((.(((........)))))))..))........(((((((.......((((((((((.....)))))))).))((((((((........)))))))).))))).))... ( -43.40) >DroSec_CAF1 17555 120 - 1 AAAUCACAAAGGAGCCGAAAACAAAUGGUGCUGGGGAUGGGGAAAGCAAAGUAACACCGCCCACCUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG ......((((((............((..((((............))))..))......((((((((((.....)))))))).))((((((((........))))))))))))))...... ( -43.50) >DroSim_CAF1 17225 120 - 1 AAAUCACAAAGGAGCCGAAAACAAAUGGUGCUGGGGAUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG ......((((((............((..((((............))))..))......((((((((((.....)))))))).))((((((((........))))))))))))))...... ( -40.80) >DroEre_CAF1 15730 114 - 1 GAAUCACAAAGGAGCCGAAAACAAAUGGUGCUGG------GGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGAAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG ......((((((............((..((((..------....))))..))......((((((((((.....)))))))).))((.(((((........))))).))))))))...... ( -35.30) >DroYak_CAF1 16459 114 - 1 AAAUCACAAAGGAGCCGAAAACAAAUGGUGCUGG------GGAAAGCGAAGUAACACCGCCCACUUUGGCUUGCUAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG ...........((((((((......(((.((.((------................))))))).)))))))).((((..((((.((((((((........)))))))).))))..)))). ( -39.69) >consensus AAAUCACAAAGGAGCCGAAAACAAAUGGUGCUGGG_AUGGGGAAAGCAAAGUAACACCGCCCACUUUGGCUUGCAAGGUGGAGCCGGAGGCGGUCCAAAACGCCUCCGCCUUUGACUGGG ......((((((............((..((((............))))..))......((((((((((.....)))))))).))((((((((........))))))))))))))...... (-38.08 = -38.36 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:46:02 2006