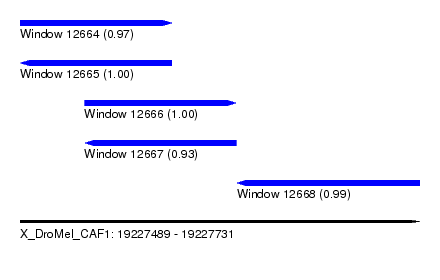
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,227,489 – 19,227,731 |
| Length | 242 |
| Max. P | 0.998258 |

| Location | 19,227,489 – 19,227,581 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 65.36 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -5.55 |
| Energy contribution | -8.63 |
| Covariance contribution | 3.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.21 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19227489 92 + 22224390 GAUAAAUGGCAAACAUGUCAUAAAAGGACUUCAUACGCAAUGCCUCGCUCGUACUCUC--------UCCCUCUCUCUCUGUAAAGAGAGAGAGAGCGAGA----------------- .....((((((....))))))......................((((((((......)--------....(((((((((....)))))))))))))))).----------------- ( -29.80) >DroSec_CAF1 14543 115 + 1 GAUAAAUGGCAAACAUGUCAUAAAAGGACUUCAUACGCAAUGCCCCACUCGCCCUCUCCCUCUCUAUCUCUCACUCUGUGGAAAGAGAGAGAG--GGAGAUGGAGCGAUGGAGCGAU .....((((((....))))))...................(((.(((.(((((((((((((((((.(((.((.(.....))).))).))))))--))))).)).))))))).))).. ( -45.60) >DroSim_CAF1 14280 74 + 1 GAUAAAUGGCAAACAUGUCAUAAAAGGACUUCAUACGCAAUGCCUC--------------------UCUCUCACUCUCUGGAAAGAGAGAGAG--G--------------------- .....((((((....)))))).....................((((--------------------((((((..((....))..)))))))))--)--------------------- ( -22.70) >DroEre_CAF1 12904 69 + 1 GAUAAAUGGCAAAUAUGUCAUAAAAGGACUUCAUACGCAUUGCCUCGCUC--------------------C------UUCGAAAGAGGGGACG----AG------------------ .....((((((....))))))......................((((..(--------------------(------(((....)))))..))----))------------------ ( -21.20) >DroYak_CAF1 13538 69 + 1 GAUAAACGGCAAACAUGUCAUAAAAGGACUUCAUACGCAAUGCCUCGCUU--------------------U----CUCCCGAAAGAGAGAAAA------------------------ .......((((.....(((.......)))...........))))....((--------------------(----(((.(....).)))))).------------------------ ( -12.79) >consensus GAUAAAUGGCAAACAUGUCAUAAAAGGACUUCAUACGCAAUGCCUCGCUC________________UC_CUC_CUCUCUGGAAAGAGAGAGAG____AG__________________ .......((((.....(((.......)))...........)))).........................(((.((((((....)))))).)))........................ ( -5.55 = -8.63 + 3.08)


| Location | 19,227,489 – 19,227,581 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 65.36 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -10.04 |
| Energy contribution | -12.52 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19227489 92 - 22224390 -----------------UCUCGCUCUCUCUCUCUUUACAGAGAGAGAGGGA--------GAGAGUACGAGCGAGGCAUUGCGUAUGAAGUCCUUUUAUGACAUGUUUGCCAUUUAUC -----------------((((.((((((((((((....)))))))))))).--------))))..........((((..((((.....(((.......))))))).))))....... ( -33.80) >DroSec_CAF1 14543 115 - 1 AUCGCUCCAUCGCUCCAUCUCC--CUCUCUCUCUUUCCACAGAGUGAGAGAUAGAGAGGGAGAGGGCGAGUGGGGCAUUGCGUAUGAAGUCCUUUUAUGACAUGUUUGCCAUUUAUC ...((((((((((.((.(((((--((((((.(((((((.....).)))))).))))))))))))))))).))))))...(((......(((.......))).....)))........ ( -50.10) >DroSim_CAF1 14280 74 - 1 ---------------------C--CUCUCUCUCUUUCCAGAGAGUGAGAGA--------------------GAGGCAUUGCGUAUGAAGUCCUUUUAUGACAUGUUUGCCAUUUAUC ---------------------.--(((((((.((((....)))).))))))--------------------).((((..((((.....(((.......))))))).))))....... ( -23.10) >DroEre_CAF1 12904 69 - 1 ------------------CU----CGUCCCCUCUUUCGAA------G--------------------GAGCGAGGCAAUGCGUAUGAAGUCCUUUUAUGACAUAUUUGCCAUUUAUC ------------------.(----((..((.((....)).------)--------------------)..)))(((((...(((((..((......))..))))))))))....... ( -15.80) >DroYak_CAF1 13538 69 - 1 ------------------------UUUUCUCUCUUUCGGGAG----A--------------------AAGCGAGGCAUUGCGUAUGAAGUCCUUUUAUGACAUGUUUGCCGUUUAUC ------------------------(((((((((....)))))----)--------------------)))...((((..((((.....(((.......))))))).))))....... ( -17.00) >consensus __________________CU____CUCUCUCUCUUUCCAGAGAG_GAG_GA________________GAGCGAGGCAUUGCGUAUGAAGUCCUUUUAUGACAUGUUUGCCAUUUAUC ........................(((((.(((((....))))).))))).......................((((.((((((.(((....))))))).))....))))....... (-10.04 = -12.52 + 2.48)


| Location | 19,227,528 – 19,227,620 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 57.06 |
| Mean single sequence MFE | -45.33 |
| Consensus MFE | -16.62 |
| Energy contribution | -20.90 |
| Covariance contribution | 4.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.28 |
| Structure conservation index | 0.37 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19227528 92 + 22224390 AUGCCUCGCUCGUACUCUC--------UCCCUCUCUCUCUGUAAAGAGAGAGAGAGCGAGA------------------GCGAGAUGGAAGGGGUACACCGUGGGUGGUCCUUUCGGU ..(((...((((..(((((--------.(.((((((((((....)))))))))).).))))------------------)))))..(((((((.(((.(....)))).)))))))))) ( -45.10) >DroSec_CAF1 14582 116 + 1 AUGCCCCACUCGCCCUCUCCCUCUCUAUCUCUCACUCUGUGGAAAGAGAGAGAG--GGAGAUGGAGCGAUGGAGCGAUGGAGCGAUGGAAGGGGUACACCGUGGGAGGUCCUUUCGGU .((((((..(((((((((((((((((.(((.((.(.....))).))).))))))--))))).)).))))....((......)).......)))))).((((.(((....)))..)))) ( -54.20) >DroEre_CAF1 12943 69 + 1 UUGCCUCGCUC--------------------C------UUCGAAAGAGGGGACG----AG-------------------GCGAGGGGGAAGGGGUACGCCGUGGGUGGUCCUUUUGGU ((((((((..(--------------------(------(((....)))))..))----))-------------------))))...(((((((.(((.(....)))).)))))))... ( -36.70) >consensus AUGCCUCGCUCG__CUCUC________UC_CUC_CUCUCUGGAAAGAGAGAGAG___GAGA__________________GCGAGAUGGAAGGGGUACACCGUGGGUGGUCCUUUCGGU ....(((((.............(((((((.(.((((((((........)))))))).).))))))).............)))))..(((((((.(((.(....)))).)))))))... (-16.62 = -20.90 + 4.28)


| Location | 19,227,528 – 19,227,620 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 57.06 |
| Mean single sequence MFE | -40.37 |
| Consensus MFE | -10.03 |
| Energy contribution | -14.87 |
| Covariance contribution | 4.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.67 |
| Structure conservation index | 0.25 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19227528 92 - 22224390 ACCGAAAGGACCACCCACGGUGUACCCCUUCCAUCUCGC------------------UCUCGCUCUCUCUCUCUUUACAGAGAGAGAGGGA--------GAGAGUACGAGCGAGGCAU .((....))..((((...))))...........((((((------------------((..(((((((((((((((......)))))))))--------))))))..))))))))... ( -45.10) >DroSec_CAF1 14582 116 - 1 ACCGAAAGGACCUCCCACGGUGUACCCCUUCCAUCGCUCCAUCGCUCCAUCGCUCCAUCUCC--CUCUCUCUCUUUCCACAGAGUGAGAGAUAGAGAGGGAGAGGGCGAGUGGGGCAU .((....))........(((((.........))))).......((((((((((.((.(((((--((((((.(((((((.....).)))))).))))))))))))))))).)))))).. ( -55.50) >DroEre_CAF1 12943 69 - 1 ACCAAAAGGACCACCCACGGCGUACCCCUUCCCCCUCGC-------------------CU----CGUCCCCUCUUUCGAA------G--------------------GAGCGAGGCAA .....((((...(((....).))...)))).......((-------------------((----((..((.((....)).------)--------------------)..)))))).. ( -20.50) >consensus ACCGAAAGGACCACCCACGGUGUACCCCUUCCAUCUCGC__________________UCUC___CUCUCUCUCUUUCCAAAGAG_GAG_GA________GAGAG__CGAGCGAGGCAU .((....))(((......)))............((((((.............(((((((((.((((((............)))))).))))))))).............))))))... (-10.03 = -14.87 + 4.84)

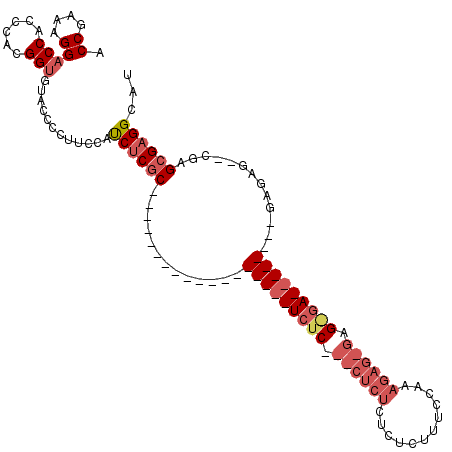
| Location | 19,227,620 – 19,227,731 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.52 |
| Mean single sequence MFE | -18.34 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.06 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
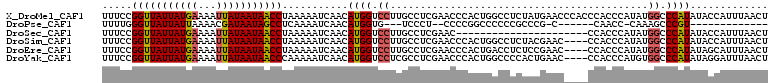
>X_DroMel_CAF1 19227620 111 - 22224390 UUUCCGGUUAUUAUGAAAAUUAUAAUAACCUAAAAAUCAACAUGGUCCUUGCCUCGAACCCACUGGCCUCUAUGAACCCACCCACCCAUAUGGCCCAUAUACCAUUUAACU .....(((((((((((...)))))))))))...........(((((...((.............((((..((((............)))).))))))...)))))...... ( -19.31) >DroPse_CAF1 52580 85 - 1 UUUUGGGUUAUUAUUAAAACGAUAAUAGCCUCAAAAUCAACAUGGUG---UCCCU--CCCCGGCCCCCCGCCCG-C------CAACC-CAAAGCCCGU------------- (((((((((((((((.....)))))))))).)))))......(((((---.....--....(((.....)))))-)------))...-..........------------- ( -19.60) >DroSec_CAF1 14698 89 - 1 UUUCCGGUUAUUAUGAAAAUUAUAAUAACCUAAAAAUCAACAUGGUCCUUGCCUCGAAC----------------------CCACCCAUAUGGCCCAUAUACCAUUUAACU .....(((((((((((...)))))))))))...........(((((....(((......----------------------..........)))......)))))...... ( -15.79) >DroSim_CAF1 14385 107 - 1 UUUCCGGUUAUUAUGAAAAUUAUAAUAACCUAAAAAUCAACAUGGUCCUUGCCUCGAACCCACUGGCCUCUACGAAC----CCACCCAUAUGGCCCAUAUACCAUUUAACU .....(((((((((((...)))))))))))...........(((((...((.............((((.........----..........))))))...)))))...... ( -17.32) >DroEre_CAF1 13012 107 - 1 UUUCCGGUUAUUAUGAAAAUUAUAAUAACCUAAAAAUCAACAUGGUCCUUGCCUCGAACCCACUGACCUCUCCGAAC----CCACCCAUAUGGCCCAUAUAGCAUUUAACU ...(((((((((((((...))))))))))).............((((..((.........))..)))).........----..........)).................. ( -14.30) >DroYak_CAF1 13643 107 - 1 UUUCCGGUUAUUAUGAAAAUUAUAAUAACCCAAAAAUCAACAUGGUCCUCGCCUCGAACCCACUGGCCCCACUGAAC----CCACCCAUGUGGCCCAUAUAGGAUUUAACU ..((((((((((((((...)))))))))))...........((((.....(((...........))).((((((...----.....)).)))).))))...)))....... ( -23.70) >consensus UUUCCGGUUAUUAUGAAAAUUAUAAUAACCUAAAAAUCAACAUGGUCCUUGCCUCGAACCCACUGGCCUCUCCGAAC____CCACCCAUAUGGCCCAUAUACCAUUUAACU .....(((((((((((...)))))))))))...........((((.((...........................................)).))))............. (-12.12 = -12.06 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:57 2006