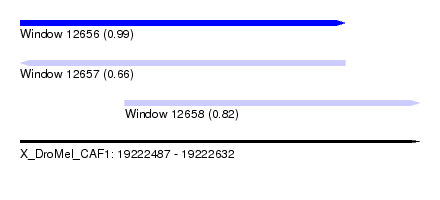
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,222,487 – 19,222,632 |
| Length | 145 |
| Max. P | 0.986423 |

| Location | 19,222,487 – 19,222,605 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19222487 118 + 22224390 UGUGUUUUCGCACCGGGCUUUUUCCGCCUGCACUUUUUCCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCG .((.(((((((..(((((.......)))))...........(((((((((((((....((((.(((((.....))))).)).)))))))).)))))))......)).)))))...)). ( -31.50) >DroSec_CAF1 9473 118 + 1 UGUGUUUUCGCACCGGGCUUUUUCCGCCUGCACUUUUUCCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCACAAGUGAAUUCCGCUGAAAAACUGCG ........((((.(((((.......))))).........................((((((.((((((..((((((.(.((.(....).)).))))))).))))))))))))..)))) ( -28.50) >DroSim_CAF1 9532 118 + 1 UGUGUUUUCGCACCGGGCUUUUUCCGCCUGCACUUUUUCCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCG .((.(((((((..(((((.......)))))...........(((((((((((((....((((.(((((.....))))).)).)))))))).)))))))......)).)))))...)). ( -31.50) >DroEre_CAF1 8234 118 + 1 UGUGUUUUCGCAUCGCGCUUUUUCCGCCUGCACUUUUUCCGCACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUUGAUGUCAGCAAAAGUGAAUUCUGCUGAAAAACUGCG ((((....)))).((((.((((((.((..............(((((((((((((....((((.(((((.....))))).).))))))))).)))))))......)).)))))).)))) ( -29.25) >DroYak_CAF1 8672 118 + 1 UGUGUUUUCGCAUCGGGCUUUUUCCGCCUGCAGUUUUUCCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGCGUUGAUGUCAGCAAAAGUGAAUUCUGCUGAAAAACUGCG ((((....))))..((((.......))))(((((((((((((((((((((((((....((((((((((.....))))))).))))))))).))))))).....))..)))))))))). ( -42.60) >consensus UGUGUUUUCGCACCGGGCUUUUUCCGCCUGCACUUUUUCCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCG .((.(((((((..(((((.......)))))...........(((((((((((((....((((.(((((.....))))).)).)))))))).)))))))......)).)))))...)). (-29.30 = -29.70 + 0.40)

| Location | 19,222,487 – 19,222,605 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19222487 118 - 22224390 CGCAGUUUUUCAGCGGAAUUCACUUUUGCUGACAUCGACUCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGUGGAAAAAGUGCAGGCGGAAAAAGCCCGGUGCGAAAACACA .(((((((((..((((((.(((.(((((.((....))...))))))))..))))))..))))))))).......((((.......(..(.(((.......)))..)..)....)))). ( -31.30) >DroSec_CAF1 9473 118 - 1 CGCAGUUUUUCAGCGGAAUUCACUUGUGCUGACAUCGACUCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGUGGAAAAAGUGCAGGCGGAAAAAGCCCGGUGCGAAAACACA .(((((((((..((((((.(((.(((.(.((....)).).)))..)))..))))))..))))))))).......((((.......(..(.(((.......)))..)..)....)))). ( -31.00) >DroSim_CAF1 9532 118 - 1 CGCAGUUUUUCAGCGGAAUUCACUUUUGCUGACAUCGACUCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGUGGAAAAAGUGCAGGCGGAAAAAGCCCGGUGCGAAAACACA .(((((((((..((((((.(((.(((((.((....))...))))))))..))))))..))))))))).......((((.......(..(.(((.......)))..)..)....)))). ( -31.30) >DroEre_CAF1 8234 118 - 1 CGCAGUUUUUCAGCAGAAUUCACUUUUGCUGACAUCAACUCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGCGGAAAAAGUGCAGGCGGAAAAAGCGCGAUGCGAAAACACA ((((.....(((((((((.....))))))))).......(((.......(((((((.(((......))).....(..(.......)..)..))))))).....)))))))........ ( -29.90) >DroYak_CAF1 8672 118 - 1 CGCAGUUUUUCAGCAGAAUUCACUUUUGCUGACAUCAACGCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGUGGAAAAACUGCAGGCGGAAAAAGCCCGAUGCGAAAACACA .((((((((((.........(((((((.((((((...(((((.((.....)).)))))......)))))))))))))..)))))))))).(((.......)))............... ( -38.80) >consensus CGCAGUUUUUCAGCGGAAUUCACUUUUGCUGACAUCGACUCGAAAUGACUUUCCGCGUGAAAAUUGUCAGAAAAGUGUGGAAAAAGUGCAGGCGGAAAAAGCCCGGUGCGAAAACACA .(((.((((((.........(((((((.(((((((((....((((....))))....)))....)))))))))))))..)))))).))).(((.......)))............... (-27.14 = -27.30 + 0.16)



| Location | 19,222,525 – 19,222,632 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19222525 107 + 22224390 CCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCGACUCUGAAUGUCGACGCUGGGAACCGA------------- .................((..((.(((....(.(((((.((((((...(((((.............))))).......)))))).))))).)..)))))..))....------------- ( -28.32) >DroSec_CAF1 9511 107 + 1 CCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCACAAGUGAAUUCCGCUGAAAAACUGCGACUCUGAAUGUCGACGCUGGGAAGCGA------------- ...(.(((..((((......))).(((....(.(((((.((((((...(((((.............))))).......)))))).))))).)..))).)..))).).------------- ( -29.22) >DroSim_CAF1 9570 107 + 1 CCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCGACUCUGAAUGUCGACGCUGGGAAGCGA------------- ...(((((((((((((....((((.(((((.....))))).)).)))))))).))))))).....((((.........((((.......)))).........)))).------------- ( -30.17) >DroEre_CAF1 8272 106 + 1 CCGCACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUUGAUGUCAGCAAAAGUGAAUUCUGCUGAAAAACUGCGACUCUGAAUGUCGACGGUGG-ACUCGA------------- ..(.((((((((((......)))...))))))))...(((((((....((((((.((.....)).))))))...((((((((.......)))).)))).)-))))))------------- ( -29.80) >DroYak_CAF1 8710 119 + 1 CCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGCGUUGAUGUCAGCAAAAGUGAAUUCUGCUGAAAAACUGCGACUCUGAAUGUCGACGGUGG-AAUCGGUGGAAUCGGUUCC ((.(((((((((((((....((((((((((.....))))))).))))))))).))))))).((((..((((...((((((((.......)))).))))..-..))))..)))).)).... ( -46.40) >consensus CCACACUUUUCUGACAAUUUUCACGCGGAAAGUCAUUUCGAGUCGAUGUCAGCAAAAGUGAAUUCCGCUGAAAAACUGCGACUCUGAAUGUCGACGCUGGGAAGCGA_____________ ...(((((((((((((....((((.(((((.....))))).)).)))))))).)))))))..................((((.......)))).((((....)))).............. (-25.64 = -25.92 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:48 2006