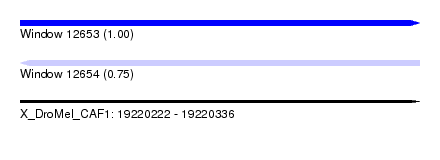
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,220,222 – 19,220,336 |
| Length | 114 |
| Max. P | 0.996971 |

| Location | 19,220,222 – 19,220,336 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -16.38 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
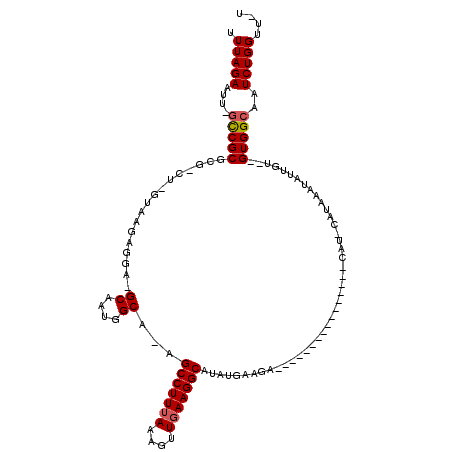
>X_DroMel_CAF1 19220222 114 + 22224390 UUUAGAAUU-GCCGCGCG-CU-GUAACAGGA-GCAAUGGCAAAGCCUUUAAAGUUGAAGGCAUAUGAAGAUAUACAUAUGUAUGUACAUACAUAAAUAUUGU--GUGGCAAUCUGGUUUU .(((((.((-((((((((-((-.(....).)-)).(((.....(((((((....))))))).((((...((((((....)))))).))))))).......))--)))))))))))).... ( -36.20) >DroSec_CAF1 7155 97 + 1 UUUAGAAUU-GCCGCGCG-CU-GUAAGAGGAGGCAAUGGCC-AGCCUUUAAAGUUGAAGGCAUAUGAAGA----------------CAUUCAUAAAUAUUGU--GUGGCAAUCUGGUU-U .(((((.((-((((((((-.(-((.......(((....)))-.(((((((....))))))).((((((..----------------..)))))).))).)))--))))))))))))..-. ( -35.60) >DroSim_CAF1 7233 86 + 1 UUUAGAAUU-GCCGCGCG-CU-GUAAGAGGA------------GCCUUUAAAGUUGAAGGCAUAUGAAGA----------------CAUUCAUAAAUAUUGU--GUGGCAAUCUGGUU-U .(((((.((-((((((((-.(-((.......------------(((((((....))))))).((((((..----------------..)))))).))).)))--))))))))))))..-. ( -31.60) >DroEre_CAF1 5915 91 + 1 UUUAGAAUUUUUCGCACG-CU-GUAAGAGGA-GCAAUGGCG-AGCCUUUAAAGUUUAAGGCA--UGAAGA----------------C----AUAAAUAUUGU--GUGGCAAUCUGGUU-U .(((((..((.(((((((-((-.(....).)-)).(((.((-.(((((........))))).--))....----------------)----)).......))--)))).)))))))..-. ( -19.10) >DroYak_CAF1 6261 94 + 1 UUUAGAAUU-GCCGCACGGCUGGCAAGAGGA-GCAAUGGCA-AGCCUUUAAAGUUGAAGGCA--UGAAGA----------------C----AUAAAUAUUGUGUGUGGCAAUCUGGUU-U .(((((.((-(((((((((((.((.......-))...))).-.(((((((....))))))).--......----------------.----..........)))))))))))))))..-. ( -31.30) >consensus UUUAGAAUU_GCCGCGCG_CU_GUAAGAGGA_GCAAUGGCA_AGCCUUUAAAGUUGAAGGCAUAUGAAGA________________CAU_CAUAAAUAUUGU__GUGGCAAUCUGGUU_U .(((((....(((((.................((....))...(((((((....)))))))...........................................)))))..))))).... (-16.38 = -16.72 + 0.34)

| Location | 19,220,222 – 19,220,336 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -7.50 |
| Energy contribution | -8.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
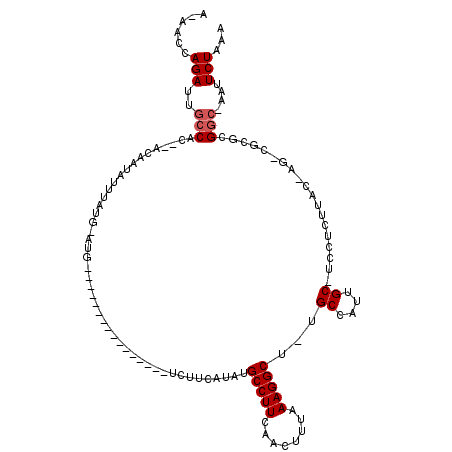
>X_DroMel_CAF1 19220222 114 - 22224390 AAAACCAGAUUGCCAC--ACAAUAUUUAUGUAUGUACAUACAUAUGUAUAUCUUCAUAUGCCUUCAACUUUAAAGGCUUUGCCAUUGC-UCCUGUUAC-AG-CGCGCGGC-AAUUCUAAA .......(((((((..--...((((.(((((((....))))))).))))..........(((((........))))).........((-.((((...)-))-.).)))))-))))..... ( -24.90) >DroSec_CAF1 7155 97 - 1 A-AACCAGAUUGCCAC--ACAAUAUUUAUGAAUG----------------UCUUCAUAUGCCUUCAACUUUAAAGGCU-GGCCAUUGCCUCCUCUUAC-AG-CGCGCGGC-AAUUCUAAA .-.....(((((((.(--.(......((((((..----------------..)))))).(((((........))))).-(((....))).........-..-.).).)))-))))..... ( -21.20) >DroSim_CAF1 7233 86 - 1 A-AACCAGAUUGCCAC--ACAAUAUUUAUGAAUG----------------UCUUCAUAUGCCUUCAACUUUAAAGGC------------UCCUCUUAC-AG-CGCGCGGC-AAUUCUAAA .-.....(((((((.(--.(......((((((..----------------..)))))).(((((........)))))------------.........-..-.).).)))-))))..... ( -18.50) >DroEre_CAF1 5915 91 - 1 A-AACCAGAUUGCCAC--ACAAUAUUUAU----G----------------UCUUCA--UGCCUUAAACUUUAAAGGCU-CGCCAUUGC-UCCUCUUAC-AG-CGUGCGAAAAAUUCUAAA .-....(((.......--...........----.----------------......--.(((((........)))))(-(((...(((-(........-))-)).)))).....)))... ( -13.80) >DroYak_CAF1 6261 94 - 1 A-AACCAGAUUGCCACACACAAUAUUUAU----G----------------UCUUCA--UGCCUUCAACUUUAAAGGCU-UGCCAUUGC-UCCUCUUGCCAGCCGUGCGGC-AAUUCUAAA .-.....(((((((.(((........(((----(----------------....))--))..............((((-.((......-.......)).))))))).)))-))))..... ( -21.42) >consensus A_AACCAGAUUGCCAC__ACAAUAUUUAUG_AUG________________UCUUCAUAUGCCUUCAACUUUAAAGGCU_UGCCAUUGC_UCCUCUUAC_AG_CGCGCGGC_AAUUCUAAA ......(((..(((.............................................(((((........)))))...((....))...................)))....)))... ( -7.50 = -8.00 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:45 2006