
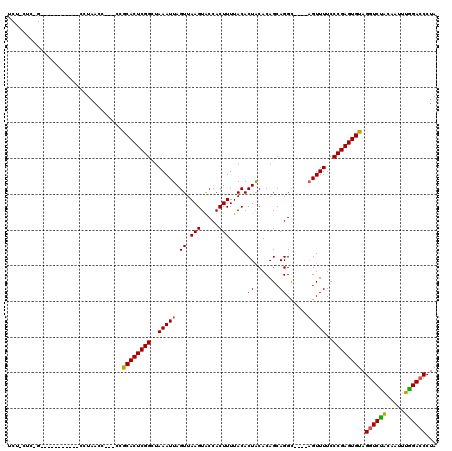
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,211,620 – 19,211,775 |
| Length | 155 |
| Max. P | 0.998518 |

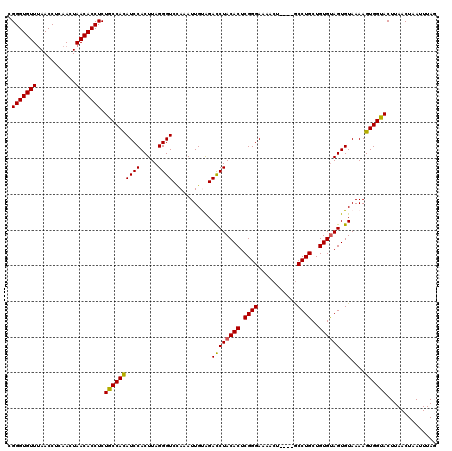
| Location | 19,211,620 – 19,211,735 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -20.26 |
| Energy contribution | -19.86 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19211620 115 + 22224390 UCUUCUCAGCCACCU-----CCUAACCGAGCAACACUCGGCUAAAUUAGUUAAGUACCACUUUUACACUACACAGCAGGCAGACAGUUUUCCCGAGUGUAGAUCUACAAUUUAGACCCUA ...............-----.....(((((.....)))))(((((((.((.(((.....)))..)).((((((....((.(((....))).))..))))))......)))))))...... ( -20.00) >DroSec_CAF1 32956 94 + 1 --------------------CCUAACCA--CCGCACUCGGCUAAAUUAGUUAAGUACCACUUUUACACUACACAGCAGGU----AGUUUUCCCGAGUGUAGGUCUACAAUUUGGACCCUA --------------------........--..((((((((........((.(((.....)))..))(((((.......))----)))....)))))))).((((((.....))))))... ( -24.10) >DroSim_CAF1 7702 94 + 1 --------------------CCUAACCA--CCGCACUCGGCUAAAUUAGUUAAGUACCACUUUUACACUGCACAGCAGGC----AGUUUUCCCGAGUGUAGGUCCACAAUUUGGACCCUA --------------------........--..((((((((..(((((.((.(((.....)))..)).((((...))))..----)))))..)))))))).((((((.....))))))... ( -29.50) >DroEre_CAF1 30086 103 + 1 UCUUCUCAG--------UCCCCUAA-----CCACACUCGGGUAAAUUAGUUAAGUGCCACUUUUACACUACACAGCAGGC----UGUUUUCCCGAGUGUAGGUCUGCAAUUUGGACCCUA ........(--------(((.(..(-----(((((((((((.(((.......((((.........))))..((((....)----))))))))))))))).)))..)......)))).... ( -28.10) >DroYak_CAF1 30908 110 + 1 UCUCCUCGGCC-CUUCCUCCCCUAA-----CCGCACUCGGGUAAAUUAGUUAAGUGCCACUUUUACACUACACAGCAGGC----AGUUUUCCCGAGUGUAGGUCUGCAAUUUGGACCCUA .......((.(-(........(..(-----(((((((((((.((((........((((...................)))----))))).))))))))).)))..)......)).))... ( -26.35) >consensus UCU_CUC_G___________CCUAACC___CCGCACUCGGCUAAAUUAGUUAAGUACCACUUUUACACUACACAGCAGGC____AGUUUUCCCGAGUGUAGGUCUACAAUUUGGACCCUA ................................((((((((..(((((.((.(((.....)))..)).((.......))......)))))..)))))))).((((((.....))))))... (-20.26 = -19.86 + -0.40)


| Location | 19,211,620 – 19,211,735 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19211620 115 - 22224390 UAGGGUCUAAAUUGUAGAUCUACACUCGGGAAAACUGUCUGCCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAGCCGAGUGUUGCUCGGUUAGG-----AGGUGGCUGAGAAGA ..(((((((.....)))))))((((((((....(((...(((((((...)))).)))..)))(((.....))).......))))))))..((((((((..-----...)))))))).... ( -32.00) >DroSec_CAF1 32956 94 - 1 UAGGGUCCAAAUUGUAGACCUACACUCGGGAAAACU----ACCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAGCCGAGUGCGG--UGGUUAGG-------------------- ((((..(((..((....((((((((.((((......----.))))..)))))).)).))..)))..)))).(((((...((((....)))--).))))).-------------------- ( -26.80) >DroSim_CAF1 7702 94 - 1 UAGGGUCCAAAUUGUGGACCUACACUCGGGAAAACU----GCCUGCUGUGCAGUGUAAAAGUGGUACUUAACUAAUUUAGCCGAGUGCGG--UGGUUAGG-------------------- ..(((((((.....))))))).(((((((....(((----(((....).)))))...(((.((((.....)))).)))..)))))))...--........-------------------- ( -29.90) >DroEre_CAF1 30086 103 - 1 UAGGGUCCAAAUUGCAGACCUACACUCGGGAAAACA----GCCUGCUGUGUAGUGUAAAAGUGGCACUUAACUAAUUUACCCGAGUGUGG-----UUAGGGGA--------CUGAGAAGA ...(((((......(.((((.(((((((((...(((----(....))))..(((((.......)))))...........)))))))))))-----)).).)))--------))....... ( -35.10) >DroYak_CAF1 30908 110 - 1 UAGGGUCCAAAUUGCAGACCUACACUCGGGAAAACU----GCCUGCUGUGUAGUGUAAAAGUGGCACUUAACUAAUUUACCCGAGUGCGG-----UUAGGGGAGGAAG-GGCCGAGGAGA ...(((((...((.(.((((..((((((((...(((----(((....).))))).....(((........)))......)))))))).))-----))...).))...)-))))....... ( -33.90) >consensus UAGGGUCCAAAUUGUAGACCUACACUCGGGAAAACU____GCCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAGCCGAGUGCGG___GGUUAGG___________C_GAG_AGA ..(((((.........))))).(((((((...........((((((...)))).))...(((........))).......)))))))................................. (-21.08 = -20.60 + -0.48)

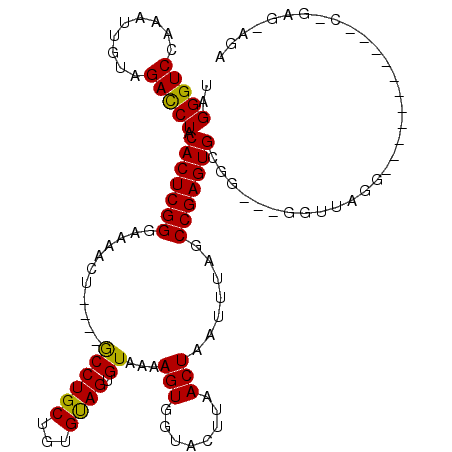

| Location | 19,211,655 – 19,211,775 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -28.88 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19211655 120 - 22224390 CGGGUGUUUAACCUCAACUAACACCUCUGCCACAUCCACUUAGGGUCUAAAUUGUAGAUCUACACUCGGGAAAACUGUCUGCCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAG .(((((((...........))))))).((((((...((((..(((((((.....)))))))((((.((((...........))))..)))))))).....)))))).............. ( -29.30) >DroSec_CAF1 32974 116 - 1 CGGGUGUUCAACCUCAACUAACACCUCUGCCACAUCCACUUAGGGUCCAAAUUGUAGACCUACACUCGGGAAAACU----ACCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAG .(((((((...........))))))).((((((((((.....))))...........((((((((.((((......----.))))..)))))).))....)))))).............. ( -29.30) >DroSim_CAF1 7720 116 - 1 CGGGUGUUUAACCUCAACUAACACCUCUGCCACAUCCACUUAGGGUCCAAAUUGUGGACCUACACUCGGGAAAACU----GCCUGCUGUGCAGUGUAAAAGUGGUACUUAACUAAUUUAG .(((((((...........))))))).((((((.(((.....(((((((.....))))))).......)))..(((----(((....).)))))......)))))).............. ( -34.70) >DroEre_CAF1 30113 116 - 1 CGGGUGUUUAACCUCAACUAACACCUCUGCCACAUCCACUUAGGGUCCAAAUUGCAGACCUACACUCGGGAAAACA----GCCUGCUGUGUAGUGUAAAAGUGGCACUUAACUAAUUUAC .(((((((...........))))))).((((((((((.....)))).....(((((...((((((.((((......----.))))..)))))))))))..)))))).............. ( -33.40) >DroYak_CAF1 30942 116 - 1 CGGGUGUUUAACCUCAACUAACACCUCUGCCAUAUCCACUUAGGGUCCAAAUUGCAGACCUACACUCGGGAAAACU----GCCUGCUGUGUAGUGUAAAAGUGGCACUUAACUAAUUUAC .(((((((...........))))))).((((((((((.....)))).....(((((...((((((.((((......----.))))..)))))))))))..)))))).............. ( -31.10) >consensus CGGGUGUUUAACCUCAACUAACACCUCUGCCACAUCCACUUAGGGUCCAAAUUGUAGACCUACACUCGGGAAAACU____GCCUGCUGUGUAGUGUAAAAGUGGUACUUAACUAAUUUAG .(((((((...........))))))).((((((((((.....))))...........((((((((.((((...........))))..)))))).))....)))))).............. (-28.88 = -28.52 + -0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:42 2006