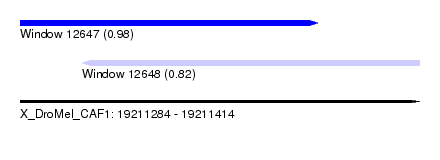
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,211,284 – 19,211,414 |
| Length | 130 |
| Max. P | 0.984744 |

| Location | 19,211,284 – 19,211,381 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.64 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -16.54 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
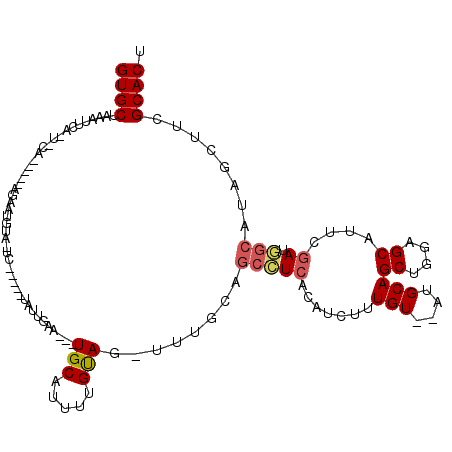
>X_DroMel_CAF1 19211284 97 + 22224390 --------------------GCAUGGUUGACAUCUUUGUAAGUGCGAAGCUAUGCCAUCGACUGCUCCAGCUGCACACACAAAGAUGUGAGGCUGCUCAUUUACAAAAUGCA---UUCAA --------------------((((..(((((((((((((..((((..((((..((........))...))))))))..))))))))))(((....))).....))).)))).---..... ( -31.30) >DroSec_CAF1 32658 98 + 1 ------------C---A-UCGCUUGCUUGACAUCUUUGUAAGUGCAAAGCUAUGCCAUCGAAUGCUCCAGCUGCAU--ACAAAGAUGUGAGGCUGCAAA-CUACAAAAUGCA---UUCAG ------------.---.-..((..((((.((((((((((..(((((..(((..((........))...))))))))--)))))))))).)))).))...-............---..... ( -30.40) >DroSim_CAF1 7414 88 + 1 --------------------------UUGACAUCUUUGUAAGUGCAAAGCUAUGCCAUCGAAUGCUCCAGCUGCAU--ACAAAGAUGUGAGGCUGCAAA-CUACAAAAUGCA---UUCAA --------------------------.(.((((((((((..(((((..(((..((........))...))))))))--)))))))))).)(..((((..-........))))---..).. ( -23.80) >DroEre_CAF1 29774 110 + 1 AGGUGGUGAUCCC---A-CCCCAUGGCUGACAUCUUUGUAAGUGCCAAGCUAUGCCAUCAAGUGCUGCAGCUGCGG--ACAAAGAAGUGAUGCUUCAAA-CUGCAAAACGCA---UUCUA .((.((((....)---)-)))).((((..(((....)))....))))((..((((..((...((((((....))))--.))..)).((..(((......-..)))..)))))---).)). ( -29.30) >DroYak_CAF1 30570 111 + 1 ------UGGUUAACCAACCCCUUUGGCUGACAUCUUUGCAAGUGCGAAGUUAUCGAAUGAGAUGCUGUGGCUGCAG--ACAAAGAAGUGAUGCUUCAAA-CUACAAAAUGCAUCAUUCAA ------..((((.((((.....)))).))))..((((((....)))))).....((((((..((((((....))))--.))..((((.....))))...-............)))))).. ( -26.90) >consensus ________________A__CCCAUGGUUGACAUCUUUGUAAGUGCAAAGCUAUGCCAUCGAAUGCUCCAGCUGCAG__ACAAAGAUGUGAGGCUGCAAA_CUACAAAAUGCA___UUCAA ...........................(.((((((((((..((((..((((..((........))...))))))))..)))))))))).).............................. (-16.54 = -17.38 + 0.84)



| Location | 19,211,304 – 19,211,414 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.93 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -8.92 |
| Energy contribution | -9.32 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19211304 110 - 22224390 GUGCUAAAUUCAUUGCAC--CCAGAAUGUAUUC-----UAUUGAA---UGCAUUUUGUAAAUGAGCAGCCUCACAUCUUUGUGUGUGCAGCUGGAGCAGUCGAUGGCAUAGCUUCGCACU ((((((...(((((((.(--(((((((((((((-----....)))---)))))))))......(((.(((.((((....)))).).)).))))).))))).))))))))........... ( -36.70) >DroSec_CAF1 32682 95 - 1 GUGCUAAAUU-------------GUAUGUA-UC-----UACUGAA---UGCAUUUUGUAG-UUUGCAGCCUCACAUCUUUGU--AUGCAGCUGGAGCAUUCGAUGGCAUAGCUUUGCACU ((((.(((..-------------.(((((.-..-----...((((---(((....(.(((-((.(((.....(((....)))--.)))))))).))))))))...)))))..))))))). ( -21.20) >DroSim_CAF1 7428 95 - 1 GUGCUAAAUU-------------GUAUGUA-UC-----UAUUGAA---UGCAUUUUGUAG-UUUGCAGCCUCACAUCUUUGU--AUGCAGCUGGAGCAUUCGAUGGCAUAGCUUUGCACU ((((.(((..-------------.((((..-.(-----(((((((---(((....(.(((-((.(((.....(((....)))--.)))))))).))))))))))))))))..))))))). ( -25.20) >DroEre_CAF1 29810 106 - 1 GUGCCAAAUCCAAUGCAC--CAAGAAUGUA-UC-----UAUAGAA---UGCGUUUUGCAG-UUUGAAGCAUCACUUCUUUGU--CCGCAGCUGCAGCACUUGAUGGCAUAGCUUGGCACU (((((((...(.((((..--((((((((((-((-----....).)---))))))((((((-(..((((.....))))..((.--...)))))))))..))))...)))).).))))))). ( -30.80) >DroYak_CAF1 30604 116 - 1 GUGCUAAAUUCACUACAUUCACAGCAUGUA-UCAACGAUAUUGAAUGAUGCAUUUUGUAG-UUUGAAGCAUCACUUCUUUGU--CUGCAGCCACAGCAUCUCAUUCGAUAACUUCGCACU ((((.........(((((.......)))))-.......(((((((((((((...((((((-...((((.....)))).....--)))))).....)))..)))))))))).....)))). ( -26.50) >consensus GUGCUAAAUUCA_U_CA_____AGAAUGUA_UC_____UAUUGAA___UGCAUUUUGUAG_UUUGCAGCCUCACAUCUUUGU__AUGCAGCUGGAGCAUUCGAUGGCAUAGCUUCGCACU ((((............................................(((.....)))........(((((.......(((....)))((....))....)).)))........)))). ( -8.92 = -9.32 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:40 2006