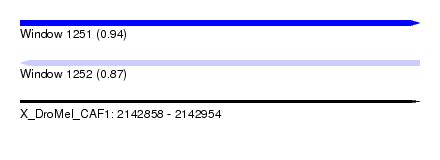
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,142,858 – 2,142,954 |
| Length | 96 |
| Max. P | 0.937996 |

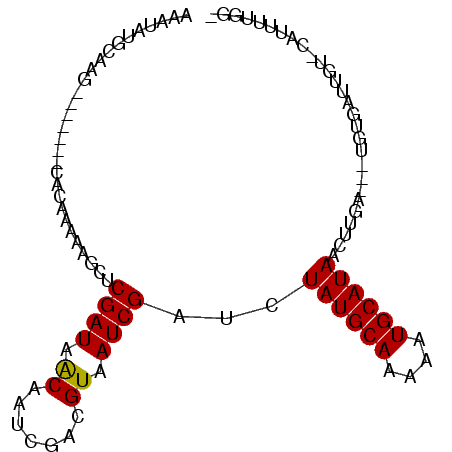
| Location | 2,142,858 – 2,142,954 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 63.21 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -8.62 |
| Energy contribution | -8.40 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.937996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2142858 96 + 22224390 AAAUAUGCAAGCGCACCACACAAAAUGCUCGAUAACAACCGAAGUAAUCGAUCUAUGCAAAAAUGCAUAUCUGAGCGAUGUGAUUGUGUAUCUCAG- ..........(((((...((((...((((((((.((.......)).)))....((((((....))))))...))))).))))..))))).......- ( -21.20) >DroEre_CAF1 24192 87 + 1 AAAUAUGCAAG-------CACAAAACGCUCGAUAACAAUCGGCGUAAUCGAUGUAUGCAAAAAUGCAUAAUGUGA---UGUGAUUGUAGAUUGUGGA .(((.(((((.-------(((...((((.((((....)))))))).((((...((((((....))))))...)))---)))).))))).)))..... ( -25.30) >DroYak_CAF1 24139 75 + 1 ------AACAG-------CAAGAAUAGCGCGAUAGCAAUUGACGUAAUCGAUCUAUGCAAAAAUGCAUAGCUUGC------CAAUG--CAUUUUGG- ------....(-------((((....(((((((....)))).))).......(((((((....))))))))))))------(((..--....))).- ( -17.50) >consensus AAAUAUGCAAG_______CACAAAAAGCUCGAUAACAAUCGACGUAAUCGAUCUAUGCAAAAAUGCAUAACUUGA___UGUGAUUGU_CAUUUUGG_ .............................((((.((.......)).))))...((((((....))))))............................ ( -8.62 = -8.40 + -0.22)

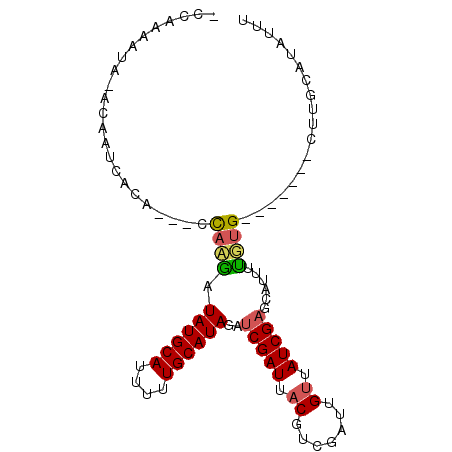
| Location | 2,142,858 – 2,142,954 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 63.21 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -8.22 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2142858 96 - 22224390 -CUGAGAUACACAAUCACAUCGCUCAGAUAUGCAUUUUUGCAUAGAUCGAUUACUUCGGUUGUUAUCGAGCAUUUUGUGUGGUGCGCUUGCAUAUUU -...(((((..(((.(.((((((.((((((((((....))))))(.(((((.((....))....))))).)..)))).)))))).).)))..))))) ( -27.10) >DroEre_CAF1 24192 87 - 1 UCCACAAUCUACAAUCACA---UCACAUUAUGCAUUUUUGCAUACAUCGAUUACGCCGAUUGUUAUCGAGCGUUUUGUG-------CUUGCAUAUUU ...........(((.((((---......((((((....))))))........((((((((....)))).))))..))))-------.)))....... ( -19.00) >DroYak_CAF1 24139 75 - 1 -CCAAAAUG--CAUUG------GCAAGCUAUGCAUUUUUGCAUAGAUCGAUUACGUCAAUUGCUAUCGCGCUAUUCUUG-------CUGUU------ -.......(--(..((------((((.(((((((....)))))))...(((...)))..))))))..))((.......)-------)....------ ( -18.80) >consensus _CCAAAAUA_ACAAUCACA___CCAAGAUAUGCAUUUUUGCAUAGAUCGAUUACGUCGAUUGUUAUCGAGCAUUUUGUG_______CUUGCAUAUUU .......................((((.((((((....))))))..(((((.((.......)).)))))......)))).................. ( -8.22 = -8.67 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:26 2006