
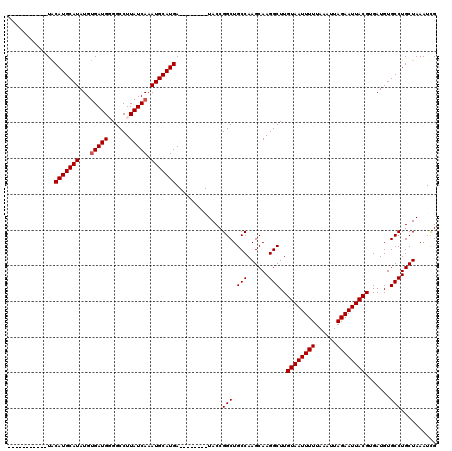
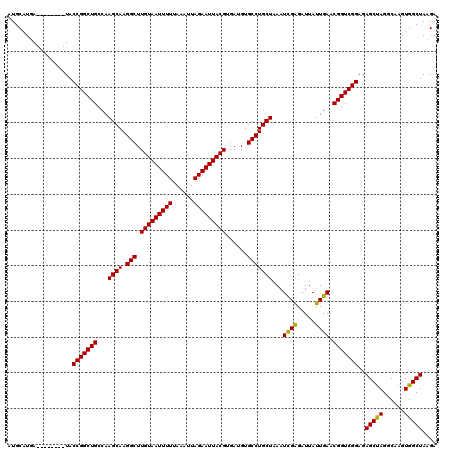
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,197,204 – 19,197,424 |
| Length | 220 |
| Max. P | 0.923327 |

| Location | 19,197,204 – 19,197,305 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.53 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19197204 101 + 22224390 -----------UACAUGCAUAUGUGAUGUGGCCUUAUCAAAUGCAUGA--------UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUUG -----------..(((((((...(((((......))))).))))))).--------....(((.(((......)))..((((((((......)))))))).......))).......... ( -27.00) >DroSec_CAF1 18720 112 + 1 UACCACAUACAUACAUGCAUAUGUGAUGUGGCCUUAUCAAAUGCAUGA--------UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCG ...((((((((((......))))).)))))(((.(((((......)))--------))..))).....((((.(((.(((((((((......)))))))))......)))))))...... ( -28.90) >DroEre_CAF1 16207 109 + 1 -----------UACAUGCAUAUGUGAUGGGGCCUUAUCAAAUGCAUGAUACCAUGAUACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUUG -----------..(((((((...((((((....)))))).)))))))...((........))......((((.(((.(((((((((......)))))))))......)))))))...... ( -27.40) >DroYak_CAF1 16093 109 + 1 -----------UGCAUGCAUAUGCGAUGGGGCCUUAUCAAAUGCAUGAUACCAUGAUACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCG -----------.(((.((((((((....(((((((........((((....))))......(((....))))))))))((((((((......)))))))))).)))))).)))....... ( -31.40) >consensus ___________UACAUGCAUAUGUGAUGGGGCCUUAUCAAAUGCAUGA________UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCG .............(((((((...(((((......))))).))))))).............(((.(((......)))..((((((((......)))))))).......))).......... (-25.27 = -25.53 + 0.25)


| Location | 19,197,204 – 19,197,305 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -23.36 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19197204 101 - 22224390 CAAUUUAGCAGGCACAUCACGUAAUUCUAAUUUAAAAAUUACAAGCCUUGCUUGGCAGCCGGUA--------UCAUGCAUUUGAUAAGGCCACAUCACAUAUGCAUGUA----------- .......((.(((.......((((((..........))))))..(((......))).))).)).--------.(((((((.((((........))))...)))))))..----------- ( -23.40) >DroSec_CAF1 18720 112 - 1 CGAUUUAGCAGGCACAUCACGUAAUUCUAAUUUAAAAAUUACAAGCCUUGCUUGGCAGCCGGUA--------UCAUGCAUUUGAUAAGGCCACAUCACAUAUGCAUGUAUGUAUGUGGUA ..........(((.......((((((..........))))))..(((......))).)))..((--------(((......)))))..(((((((.((((((....))))))))))))). ( -30.90) >DroEre_CAF1 16207 109 - 1 CAAUUUAGCAGGCACAUCACGUAAUUCUAAUUUAAAAAUUACAAGCCUUGCUUGGCAGCCGGUAUCAUGGUAUCAUGCAUUUGAUAAGGCCCCAUCACAUAUGCAUGUA----------- ..........(((.......((((((..........))))))..(((......))).)))(((((....)))))((((((.((((........))))...))))))...----------- ( -24.10) >DroYak_CAF1 16093 109 - 1 CGAUUUAGCAGGCACAUCACGUAAUUCUAAUUUAAAAAUUACAAGCCUUGCUUGGCAGCCGGUAUCAUGGUAUCAUGCAUUUGAUAAGGCCCCAUCGCAUAUGCAUGCA----------- .......((.(((.......((((((..........))))))..(((......))).)))(((((....)))))((((((.((....(....)....)).)))))))).----------- ( -25.90) >consensus CAAUUUAGCAGGCACAUCACGUAAUUCUAAUUUAAAAAUUACAAGCCUUGCUUGGCAGCCGGUA________UCAUGCAUUUGAUAAGGCCACAUCACAUAUGCAUGUA___________ .......((.(((.......((((((..........))))))..(((......))).))).))..........(((((((.((((........))))...)))))))............. (-23.36 = -23.18 + -0.19)

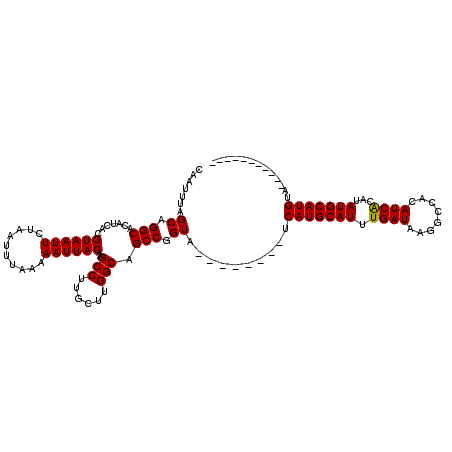

| Location | 19,197,233 – 19,197,345 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -30.05 |
| Energy contribution | -29.43 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19197233 112 + 22224390 AUGCAUGA--------UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUUGAGAUUAUUGAACGGUCGGAGAGCCAGGCAAGUGGCUAAGA ........--------..(((((((...((((.(((.(((((((((......)))))))))......)))))))...(..(.....)..).)))))))..(((((......))))).... ( -30.60) >DroSec_CAF1 18760 112 + 1 AUGCAUGA--------UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCGAGUUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUACGA ........--------..(((((((...((((.(((.(((((((((......)))))))))......)))))))...((((.....)))).)))))))..(((((......))))).... ( -30.50) >DroEre_CAF1 16236 120 + 1 AUGCAUGAUACCAUGAUACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUUGGGAUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUACGA ...((((....))))...((((((((((((((.(((.(((((((((......)))))))))......)))))))....)))..........)))))))..(((((......))))).... ( -30.90) >DroYak_CAF1 16122 120 + 1 AUGCAUGAUACCAUGAUACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCGAGAUUAUUGAACGGUCGGAGAGCUAGCCAAGUGGCUAAGA ...((((....))))...(((((((...((((.(((.(((((((((......)))))))))......)))))))...((((.....)))).))))))).....(((((....)))))... ( -35.70) >consensus AUGCAUGA________UACCGGCUGCCAAGCAAGGCUUGUAAUUUUUAAAUUAGAAUUACGUGAUGUGCCUGCUAAAUCGAGAUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUAAGA ..................(((((((...((((.(((.(((((((((......)))))))))......)))))))...((((.....)))).)))))))..(((((......))))).... (-30.05 = -29.43 + -0.62)


| Location | 19,197,305 – 19,197,424 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -28.15 |
| Energy contribution | -28.78 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19197305 119 + 22224390 AGAUUAUUGAACGGUCGGAGAGCCAGGCAAGUGGCUAAGAAAAUGUUGUUUAGAACCAGGUCGAGAAGUAUGGGACAUUCUGGGCCAGAGCAACA-GCUUUCCAGCCACGAAAACAUUGC ............(((.(((((((..(((.....))).......((((((((.(..(((((((...........)))...))))..).))))))))-))))))).)))............. ( -33.90) >DroSec_CAF1 18832 118 + 1 AGUUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUACGAAAAUGUUGUUUAGAACCAGGUCGAGAAGUAUGAGACGUUCUGGGCCAGAGCAACA-GCUUUCCAGCCACGAAAACAUUG- .((((.(((...(((.((((((((.(((.....)))........(((((((.(..((((..((............))..))))..).))))))))-))))))).))).)))))))....- ( -36.40) >DroEre_CAF1 16316 119 + 1 GGAUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUACGAAAAUGUUGUUUAGAGCCAGGUCGAGAAGUAUGGGACUUAC-GGGCCAGAGCAACAAGCUUUCCUGCCACGGAAACACUUC ............(((.((((((((.(((.....))).......((((((((.(.(((.((((...........))))...-.)))).)))))))))))))))).)))..(....)..... ( -37.60) >DroYak_CAF1 16202 119 + 1 AGAUUAUUGAACGGUCGGAGAGCUAGCCAAGUGGCUAAGAAAAUGUUGUUUAGAACCAGGUCGAGAAGUAUGGCACUUUC-GGGCCAAAGCAACAAGCUUUCUUGCCACGAAAACAUUUC ............(((.((((((((((((....)))))......((((((((.......((((..(((((.....))))).-.)))).)))))))).))))))).)))............. ( -34.60) >consensus AGAUUAUUGAACGGUCGGAGAGCUAGGCAAGUGGCUAAGAAAAUGUUGUUUAGAACCAGGUCGAGAAGUAUGGGACUUUC_GGGCCAGAGCAACA_GCUUUCCAGCCACGAAAACAUUGC ............(((.((((((((((.(....).)))......((((((((.(..(((((((...........)))...))))..).)))))))).))))))).)))............. (-28.15 = -28.78 + 0.63)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:23 2006