| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,122,254 – 2,122,367 |
| Length | 113 |
| Max. P | 0.740855 |

| Location | 2,122,254 – 2,122,367 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -25.91 |
| Energy contribution | -25.50 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
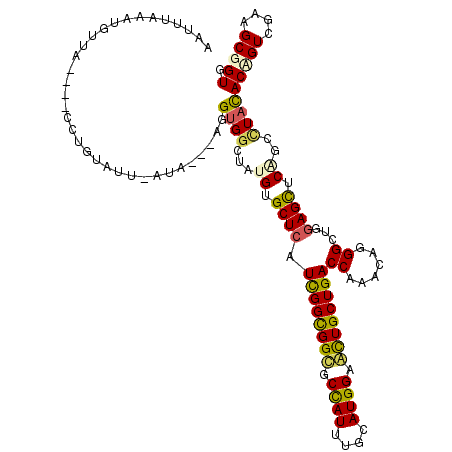
>X_DroMel_CAF1 2122254 113 - 22224390 UAUUUAAAUGAUU----CUUGUGUUUACA---AGGCGGCUAUGUGCUCAUCGGUGGUGCCAUCUGCAUGGAAUUGCUGACCAAACAGGGCUGGAGCUCGGCAUAUACCGUGGAAGCGGUU .............----(((((....)))---))((((.((((((((..((((..((.((((....)))).))..)))).......((((....)))))))))))))))).......... ( -34.60) >DroVir_CAF1 3896 107 - 1 GAUUUAAAAAUCAUAAUC----------C---AGGUGGCUAUGUGCUGAUCGGCGGUGCCAUUUGCAUGGAACUGCUGACCAAACAAGGCUGGAGUUCCGCCUACACAGUCGAAGCCGUG ((((....))))......----------.---....((((.((.((((.((((((((.((((....)))).)))))))).......((((.((....))))))...)))))).))))... ( -35.30) >DroPse_CAF1 3583 116 - 1 AAUUUUAAUUUGA----UCUGUAUUAAUAUGUAGGUGGAUAUGUGCUCAUCGGCGGCGCCAUUUGCAUGGAGCUGCUGACCAAACAGGGCUGGAGCUCAGCCUACACUGUCGAGGCGGUA .............----.((((.(..((((((((((.....((.((((.((((((((.((((....)))).))))))))((.....))....)))).))))))))).)))..).)))).. ( -39.70) >DroMoj_CAF1 4095 117 - 1 GAUUCUAAUGCUUUCAUCGUUUCUUCAUU---AGGUGGCUAUGUGCUCAUUGGUGGCGCUAUUUGCAUGGAGCUGCUGACCAAGCAAGGCUGGAGUUCCGCCUACACAGUCGAAGCCGUG ...(((((((...............))))---))).((((...((((..(..(..((.((((....)))).))..)..)...))))((((.((....))))))..........))))... ( -31.46) >DroAna_CAF1 3754 110 - 1 UAAAUAGAAGAUU----CUUGUUU---CC---AGGUGGCUAUGUGCUCAUCGGUGGAGCCAUCUGCAUGGAACUGCUGACCAAACAGGGUUGGAGCUCAGCUUAUACCGUCGAGGCGGUG .............----...((((---((---((((((((...((((....)))).)))))))))...))))).(((((((((......))))...)))))...((((((....)))))) ( -33.70) >DroPer_CAF1 3587 116 - 1 AAUUUUAAUUUGA----UCUGUAUUAAUAUAUAGGUGGAUAUGUGCUCAUCGGCGGCGCCAUUUGCAUGGAGCUGCUGACCAAACAGGGCUGGAGCUCAGCCUACACUGUCGAGGCGGUA .........((.(----(((((((....)))))))).))..(((.....((((((((.((((....)))).))))))))....)))((((((.....))))))..(((((....))))). ( -39.90) >consensus AAUUUAAAUGUUA____CCUGUAUU_AUA___AGGUGGCUAUGUGCUCAUCGGCGGCGCCAUUUGCAUGGAACUGCUGACCAAACAGGGCUGGAGCUCAGCCUACACAGUCGAAGCGGUG ..................................((((...((.((((.((((((((.((((....)))).))))))))((......))...)))).))..))))(((((....))))). (-25.91 = -25.50 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:25 2006