
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,175,767 – 19,175,857 |
| Length | 90 |
| Max. P | 0.646290 |

| Location | 19,175,767 – 19,175,857 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
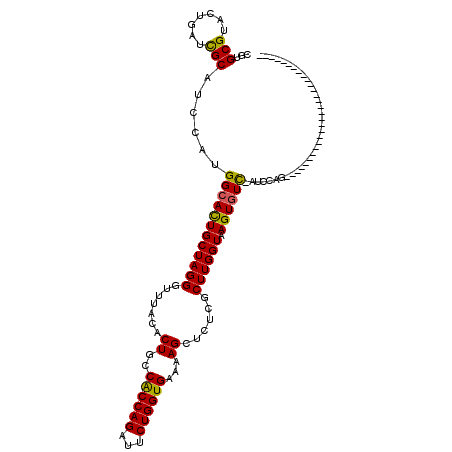
>X_DroMel_CAF1 19175767 90 + 22224390 -----------------------------CUGGAC-GACACUUACCAAGCGAAAGCUUUUCACCAGAAUCUGGUGGCAGUGUAAACCCUAGCAGUGCCAUGGAGGCGAUCAGUACGCACG -----------------------------((((.(-(...(((...((((....))))...(((((...)))))((((.(((........))).))))...))).)).))))........ ( -26.90) >DroVir_CAF1 12785 120 + 1 GGUUUCUGGUUUCUGGAUUUGAGUGUGUAUUAGAUGGACACUUACCAAGCGAGAGCUUCUCACCAGAAUCUGGUGGCAGGGUAAACCCUAGCAGUGCCAUCGAUGCGAUCAAUACGCAUG ....((..(...)..)).....(((((((((.(((((.((((....((((....))))..((((((...))))))((((((....)))).)))))))))))(((...)))))))))))). ( -42.30) >DroGri_CAF1 12416 117 + 1 GGUUUUUGGAUUGU---UUCUGCUGUGUAUUAAAUGGACACUUACCAAGCGAGAGCUUUUCACCAGAAUCUGGUGGCAGUGUGAACCCUAGCAGUGCCAUCGAUGCGAUCAAUACGCACG ((((((((((..((---(..((((..(((.............)))..))))..)))..))))..))))))((((((((.(((........))).)))))))).((((.......)))).. ( -33.12) >DroWil_CAF1 16446 88 + 1 -----------------------------CUG-AU-GA-ACUUACCAAGCGAGAGCUUCUCACCAGAAUCUGGUGGCAGUGUAAAACCUAGCAAUGCCAUCGAUGCGAUCAAUACGCAUG -----------------------------(((-.(-((-.......((((....)))).))).)))....((((((((.(((........))).))))))))(((((.......))))). ( -25.20) >DroYak_CAF1 16912 90 + 1 -----------------------------CUGGAC-GACACUUACCAAGCGAAAGCUUUUCACCAGAAUCUGGUGGCAGUGUAAACCCUAGCAGUGCCAUGGAGGCGAUCAGUACGCACG -----------------------------((((.(-(...(((...((((....))))...(((((...)))))((((.(((........))).))))...))).)).))))........ ( -26.90) >DroPer_CAF1 43229 90 + 1 -----------------------------CUGGGC-AACACUUACCAAGUGAAAGCUUCUCACCAGAAUCUGGCGGAAGUGUAAACCCUAGCAGUGCCAUGGAGGCAAUUAGUACGCACG -----------------------------(((((.-.((((((.((((((....))))....((((...)))).))))))))....)))))...((((.....))))............. ( -24.50) >consensus _____________________________CUGGAC_GACACUUACCAAGCGAAAGCUUCUCACCAGAAUCUGGUGGCAGUGUAAACCCUAGCAGUGCCAUCGAGGCGAUCAAUACGCACG .............................((((....(((((....((((....)))).(((((((...))))))).))))).....))))..((((....((.....)).....)))). (-19.28 = -19.37 + 0.09)



| Location | 19,175,767 – 19,175,857 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -28.18 |
| Consensus MFE | -20.56 |
| Energy contribution | -20.17 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19175767 90 - 22224390 CGUGCGUACUGAUCGCCUCCAUGGCACUGCUAGGGUUUACACUGCCACCAGAUUCUGGUGAAAAGCUUUCGCUUGGUAAGUGUC-GUCCAG----------------------------- .(..((((((.((((((.....)))...((.(((((((.......((((((...))))))..))))))).))..))).)))).)-)..)..----------------------------- ( -24.90) >DroVir_CAF1 12785 120 - 1 CAUGCGUAUUGAUCGCAUCGAUGGCACUGCUAGGGUUUACCCUGCCACCAGAUUCUGGUGAGAAGCUCUCGCUUGGUAAGUGUCCAUCUAAUACACACUCAAAUCCAGAAACCAGAAACC .(((((.......))))).(((((((((((.((((....))))))((((((...))))))..((((....))))....)))).)))))................................ ( -34.20) >DroGri_CAF1 12416 117 - 1 CGUGCGUAUUGAUCGCAUCGAUGGCACUGCUAGGGUUCACACUGCCACCAGAUUCUGGUGAAAAGCUCUCGCUUGGUAAGUGUCCAUUUAAUACACAGCAGAA---ACAAUCCAAAAACC .((((.(((((((...))))))))))).....(((((....((((((((((...))))))..((((....)))).....((((.........)))).))))..---..)))))....... ( -31.40) >DroWil_CAF1 16446 88 - 1 CAUGCGUAUUGAUCGCAUCGAUGGCAUUGCUAGGUUUUACACUGCCACCAGAUUCUGGUGAGAAGCUCUCGCUUGGUAAGU-UC-AU-CAG----------------------------- .(((((.......))))).(((((..(((((((((......((.(((((((...)))))).).)).....)))))))))..-))-))-)..----------------------------- ( -26.90) >DroYak_CAF1 16912 90 - 1 CGUGCGUACUGAUCGCCUCCAUGGCACUGCUAGGGUUUACACUGCCACCAGAUUCUGGUGAAAAGCUUUCGCUUGGUAAGUGUC-GUCCAG----------------------------- .(..((((((.((((((.....)))...((.(((((((.......((((((...))))))..))))))).))..))).)))).)-)..)..----------------------------- ( -24.90) >DroPer_CAF1 43229 90 - 1 CGUGCGUACUAAUUGCCUCCAUGGCACUGCUAGGGUUUACACUUCCGCCAGAUUCUGGUGAGAAGCUUUCACUUGGUAAGUGUU-GCCCAG----------------------------- .............((((.....))))......((((..(((((((((((((...)))))((((...))))....)).)))))).-))))..----------------------------- ( -26.80) >consensus CGUGCGUACUGAUCGCAUCCAUGGCACUGCUAGGGUUUACACUGCCACCAGAUUCUGGUGAAAAGCUCUCGCUUGGUAAGUGUC_AUCCAG_____________________________ ...(((.......)))......((((((((((((.......((..((((((...))))))...))......)))))).)))))).................................... (-20.56 = -20.17 + -0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:45:13 2006