
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,143,458 – 19,143,603 |
| Length | 145 |
| Max. P | 0.850711 |

| Location | 19,143,458 – 19,143,563 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.22 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -24.20 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
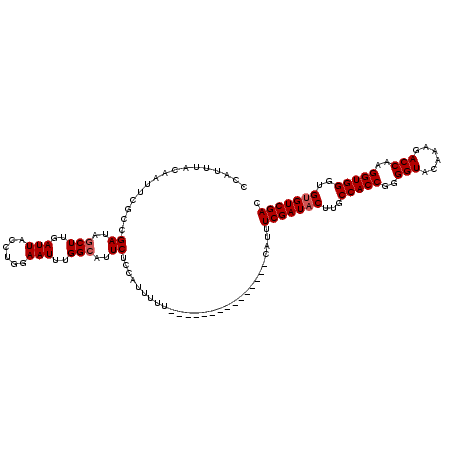
>X_DroMel_CAF1 19143458 105 + 22224390 CCAUUUACAAUUCGCCGAUAGCUUGAUUACCUGGAAUUUGGCAUUCUCCAUUUUU---------------CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGAA .............(((((...((.(.....).))...))))).............---------------...((((((((...(((((..(((......)))..)))))..)))))))) ( -30.50) >DroSim_CAF1 13059 105 + 1 CCAUUUACAAUGCGGCGAUAGCUUGAUUACCUGGAAUUUGGCAUUCUCCAUUUUU---------------CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGAC .......(((.((.......)))))......(((((..(((......)))..)))---------------))..(((((((...(((((..(((......)))..)))))..))))))). ( -31.20) >DroYak_CAF1 8019 120 + 1 CCAUUUACAAUUCGUCGAUAUCUUGAUUACCUGGAAUUGGGCAUUCUUCAUUCUUCAUUUUUCCUUUUUUUAUUUCGAUUCUUGCCACCCGGGUACAAAGACCAAGGUGGGUGUGUCGAC .............(((((((((((...(((((((.....((((....((...........................))....))))..)))))))..)))(((......))))))))))) ( -26.23) >consensus CCAUUUACAAUUCGCCGAUAGCUUGAUUACCUGGAAUUUGGCAUUCUCCAUUUUU_______________CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGAC ................((..(((..(((......)))..)))..))............................(((((((...(((((..(((......)))..)))))..))))))). (-24.20 = -24.87 + 0.67)


| Location | 19,143,498 – 19,143,603 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -28.05 |
| Energy contribution | -28.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
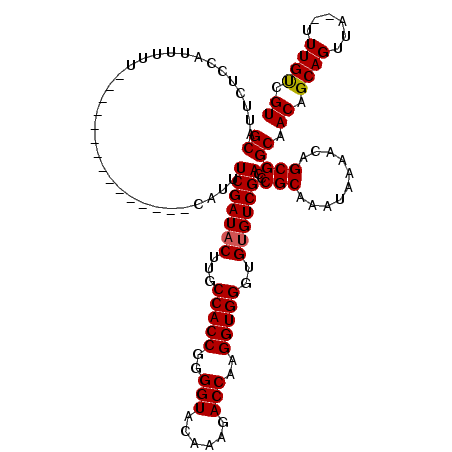
>X_DroMel_CAF1 19143498 105 + 22224390 GCAUUCUCCAUUUUU---------------CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGAACCGCAAAUAAAACAGCGGCAACAGCAGUUGUGUUUGUCGU ...............---------------...((((((((...(((((..(((......)))..)))))..)))))))).((((((((.(((.(((....).)).))).)))))).)). ( -34.40) >DroSim_CAF1 13099 103 + 1 GCAUUCUCCAUUUUU---------------CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGACCCGCAAAUAAAACAGCGGCAACAGCAGUUA--CUUGUCGU ((.............---------------....(((((((...(((((..(((......)))..)))))..))))))).((((..........)))).....)).....--........ ( -29.90) >DroYak_CAF1 8059 118 + 1 GCAUUCUUCAUUCUUCAUUUUUCCUUUUUUUAUUUCGAUUCUUGCCACCCGGGUACAAAGACCAAGGUGGGUGUGUCGACACGCAAAUAGAAGAGCGGCAACAGCAGUUU--UUUGCGGU ((.((((((...................................(((((..(((......)))..)))))(((((....))))).....))))))..))....((((...--.))))... ( -31.00) >consensus GCAUUCUCCAUUUUU_______________CAUUUCGAUACUUGCCACCGGGGUACAAAGACCAAGGUGGGUGUGUCGACCCGCAAAUAAAACAGCGGCAACAGCAGUUA__UUUGUCGU ((................................(((((((...(((((..(((......)))..)))))..)))))))..(((..........))))).((.((((......)))).)) (-28.05 = -28.17 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:59 2006