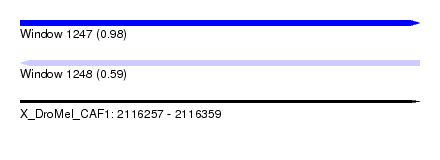
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,116,257 – 2,116,359 |
| Length | 102 |
| Max. P | 0.980570 |

| Location | 2,116,257 – 2,116,359 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -15.04 |
| Energy contribution | -15.92 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
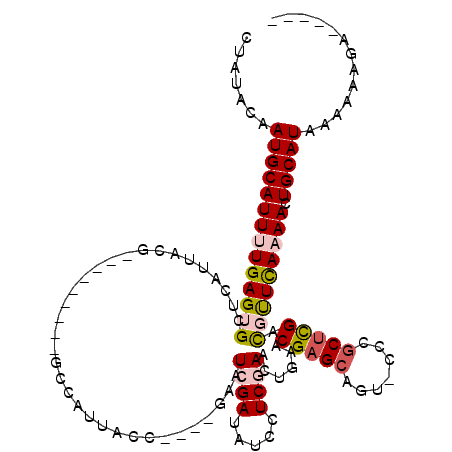
>X_DroMel_CAF1 2116257 102 + 22224390 CCAUACAAUGCAUUUUGAGUGCUCAUUACG---------GCCAUUACC----GAAUCGAUAUUCUCGAAAUGAACGAGCCGAACUGGCUCGACGUUCAAAACUGCAUAAAAAAGG----- .......((((((((((((((.(((((.((---------(......))----)..((((.....))))))))).((((((.....)))))).))))))))).)))))........----- ( -29.70) >DroSec_CAF1 4391 104 + 1 CUAUACAAUGCAUUUUGAGUGCUCAUUACG---------GCCAUUACCGACCGAAUCGAUAUCCUCGAACUGAACGAGCAGU-CCCGCUCGACUUUCA-AACUGCAUAAAAAAGA----- .......(((((.(((((((((((....((---------(......)))......((((.....)))).......)))))((-(......))).))))-)).)))))........----- ( -25.00) >DroSim_CAF1 4384 103 + 1 CUAUACAAUGCAUUUUGAGUGCUCAUUACG---------GCCAUUACCGACCGAAUCGAUAUCCUCGAACUGAACGAGCAGU-CCCGCUCGACUUUCA-AACUGCAUAAA-AAGA----- .......(((((.(((((((((((....((---------(......)))......((((.....)))).......)))))((-(......))).))))-)).)))))...-....----- ( -25.00) >DroEre_CAF1 4327 113 + 1 CUAUACAAUGCAUUUUGAGUGCUCAUUACGGCCAUUACGGCCAUUACC----AAAUCGAUAUCCUCAAACUGAACGAGGAA---CUGCUCGAUGCUCAAAAAUGCAUAAAAUGCAAAAGC .......(((((((((((((.........((((.....))))......----..(((((..(((((.........))))).---....))))))))).)))))))))............. ( -31.00) >DroYak_CAF1 4429 103 + 1 CUAUACAAUGCAUUUUGAGUGCUCAUUACG---------GCCAUUACC----AAAUCGAUAUCCUCAAACCGAACGAGCAGU-UCCGCCUGAUGUUUAAAAAUGCAUAAAAAAAAAA--- .......(((((((((.......(((((.(---------((.......----.....((.....)).....((((.....))-)).))))))))....)))))))))..........--- ( -19.30) >consensus CUAUACAAUGCAUUUUGAGUGCUCAUUACG_________GCCAUUACC____GAAUCGAUAUCCUCGAACUGAACGAGCAGU_CCCGCUCGACGUUCAAAACUGCAUAAAAAAGA_____ .......((((((((((((((..................................((((.....))))......(((((.......))))).))))))))).)))))............. (-15.04 = -15.92 + 0.88)

| Location | 2,116,257 – 2,116,359 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.82 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
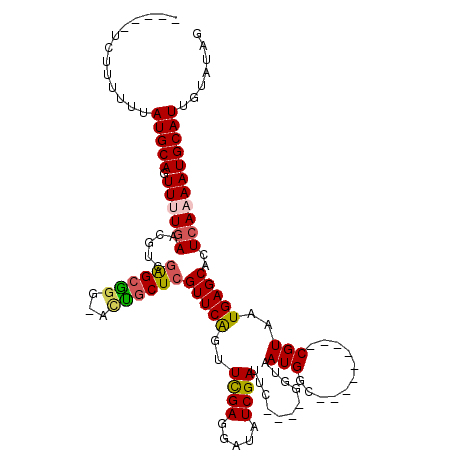
>X_DroMel_CAF1 2116257 102 - 22224390 -----CCUUUUUUAUGCAGUUUUGAACGUCGAGCCAGUUCGGCUCGUUCAUUUCGAGAAUAUCGAUUC----GGUAAUGGC---------CGUAAUGAGCACUCAAAAUGCAUUGUAUGG -----((......(((((.((((((.....(((((.....)))))(((((((((((.....))))..(----(((....))---------)).)))))))..))))))))))).....)) ( -32.70) >DroSec_CAF1 4391 104 - 1 -----UCUUUUUUAUGCAGUU-UGAAAGUCGAGCGGG-ACUGCUCGUUCAGUUCGAGGAUAUCGAUUCGGUCGGUAAUGGC---------CGUAAUGAGCACUCAAAAUGCAUUGUAUAG -----........(((((.((-(((.....(((((..-..)))))(((((..((((.....))))..((((((....))))---------))...)))))..))))).)))))....... ( -31.40) >DroSim_CAF1 4384 103 - 1 -----UCUU-UUUAUGCAGUU-UGAAAGUCGAGCGGG-ACUGCUCGUUCAGUUCGAGGAUAUCGAUUCGGUCGGUAAUGGC---------CGUAAUGAGCACUCAAAAUGCAUUGUAUAG -----....-...(((((.((-(((.....(((((..-..)))))(((((..((((.....))))..((((((....))))---------))...)))))..))))).)))))....... ( -31.40) >DroEre_CAF1 4327 113 - 1 GCUUUUGCAUUUUAUGCAUUUUUGAGCAUCGAGCAG---UUCCUCGUUCAGUUUGAGGAUAUCGAUUU----GGUAAUGGCCGUAAUGGCCGUAAUGAGCACUCAAAAUGCAUUGUAUAG ((....)).....(((((((((.(((.(((((....---.((((((.......))))))..)))))((----.((.((((((.....)))))).)).))..))))))))))))....... ( -33.80) >DroYak_CAF1 4429 103 - 1 ---UUUUUUUUUUAUGCAUUUUUAAACAUCAGGCGGA-ACUGCUCGUUCGGUUUGAGGAUAUCGAUUU----GGUAAUGGC---------CGUAAUGAGCACUCAAAAUGCAUUGUAUAG ---..........(((((((((......((((((.((-((.....)))).)))))).((..(((...(----(((....))---------))...)))....)))))))))))....... ( -23.10) >consensus _____UCUUUUUUAUGCAGUUUUGAACGUCGAGCGGG_ACUGCUCGUUCAGUUCGAGGAUAUCGAUUC____GGUAAUGGC_________CGUAAUGAGCACUCAAAAUGCAUUGUAUAG .............(((((.((((((.....((((((...))))))(((((..((((.....))))...........(((...........)))..)))))..)))))))))))....... (-17.74 = -17.82 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:23 2006