
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,112,480 – 19,112,586 |
| Length | 106 |
| Max. P | 0.999232 |

| Location | 19,112,480 – 19,112,586 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -19.01 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.69 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
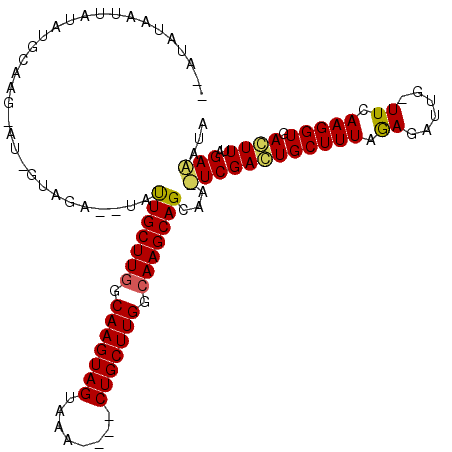
>X_DroMel_CAF1 19112480 106 + 22224390 --AUAUGAUUAUAUGCAAGCAUCGUAGA--UAUUGCUUGGCAAGUAGUAAA----CUGCUUGACAAGCAGCAAGUCGACUGCUUUAGAGAUUG-UUCAAGGUGACUUUAGAUAUA --..(..(((...((.((((((((....--..(((((((.(((((((....----))))))).))))))).....))).)))))))..)))..-)..(((....)))........ ( -28.60) >DroSec_CAF1 1127 97 + 1 --AUAUAAUAUGAU---------UUAGA--UAUUGCUUGGCAAGUAGUAAA----CUGCUUGGCAAGCAGCAAUUCGAGUGCUUUAGAGAUUG-UUCAAGGUGACUUUAGAAAUA --..(((((....(---------(((((--...((((((.(((((((....----))))))).))))))(((.......))))))))).))))-)..(((....)))........ ( -26.60) >DroYak_CAF1 1076 113 + 1 AUACAUAUAUAUUUGCAAGAAUUG-AAAUGCACUGCUUAGCAAGUAGUAAACUGACUGCUUGGCAAGCAGCAA-UCGAUUGCUUUAAAGAUUGCUUCAAGGUGAGUUUAGAAAUG ...(((.((.(((..(.....(((-((......(((((..((((((((......))))))))..)))))((((-((............))))))))))).)..))).))...))) ( -28.00) >consensus __AUAUAAUUAUAUGCAAG_AU_GUAGA__UAUUGCUUGGCAAGUAGUAAA____CUGCUUGGCAAGCAGCAA_UCGACUGCUUUAGAGAUUG_UUCAAGGUGACUUUAGAAAUA ................................(((((((.(((((((........))))))).)))))))...((((((((((((.((......)).))))).))))..)))... (-19.01 = -19.13 + 0.12)


| Location | 19,112,480 – 19,112,586 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -13.43 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
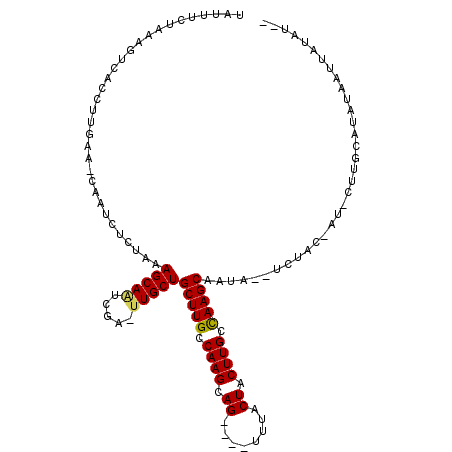
>X_DroMel_CAF1 19112480 106 - 22224390 UAUAUCUAAAGUCACCUUGAA-CAAUCUCUAAAGCAGUCGACUUGCUGCUUGUCAAGCAG----UUUACUACUUGCCAAGCAAUA--UCUACGAUGCUUGCAUAUAAUCAUAU-- .....................-.........(((((.(((...((.((((((.((((.((----....)).)))).)))))).))--....))))))))..............-- ( -20.30) >DroSec_CAF1 1127 97 - 1 UAUUUCUAAAGUCACCUUGAA-CAAUCUCUAAAGCACUCGAAUUGCUGCUUGCCAAGCAG----UUUACUACUUGCCAAGCAAUA--UCUAA---------AUCAUAUUAUAU-- .................(((.-..........((((.......))))(((((.((((.((----....)).)))).)))))....--.....---------.)))........-- ( -15.40) >DroYak_CAF1 1076 113 - 1 CAUUUCUAAACUCACCUUGAAGCAAUCUUUAAAGCAAUCGA-UUGCUGCUUGCCAAGCAGUCAGUUUACUACUUGCUAAGCAGUGCAUUU-CAAUUCUUGCAAAUAUAUAUGUAU ................(((((((((((............))-))))(((((((..(((((..((....))..)))))..)))).))).))-)))....((((........)))). ( -21.30) >consensus UAUUUCUAAAGUCACCUUGAA_CAAUCUCUAAAGCAAUCGA_UUGCUGCUUGCCAAGCAG____UUUACUACUUGCCAAGCAAUA__UCUAC_AU_CUUGCAUAUAAUUAUAU__ ................................(((((.....)))))(((((.((((.((........)).)))).))))).................................. (-13.43 = -13.43 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:48 2006