
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,105,257 – 19,105,377 |
| Length | 120 |
| Max. P | 0.671829 |

| Location | 19,105,257 – 19,105,377 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -44.62 |
| Consensus MFE | -30.47 |
| Energy contribution | -29.75 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19105257 120 + 22224390 UCGCCACCGCUCUCUACGACUACGAACCAAAGGCGGGCAGCAAUCAGCUGCAGCUGCGCACUGACUGCCAGGUGCUGGUCAUUGGCAAGGAUGGGGACAGCAAGGGCUGGUGGCGUGGCA .((((((((((((....(((((.........(((..(((((.....))))).)))((((.(((.....))))))))))))...........((....))...))))).)))))))..... ( -48.70) >DroPse_CAF1 2015 120 + 1 UAGCCACUGCCCUGUACGAUUACGAACCGAAGCCAGGGAGCAAUCAAUUGCAGCUGCGCACCGACUGCCAGAUAUUGGUUAUUGGCAAAGAUGGGGACAGCAAGGGCUGGUGGCGAGGAA ..(((((((((((((...........((((((((((...((((....))))..(((.(((.....))))))...)))))).))))......((....)))).))))).))))))...... ( -41.10) >DroGri_CAF1 3824 120 + 1 UCGCCACCGCCCUAUACGACUUUGAUCCGAAGGCGGGCAGCAAUCAGCUGCAACUGCGUACCGAUUGCCAGGUACUUGUAAUUGGCAAGGAUGGCGACAGCAAAGGCUGGUGGCGUGGUA .((((((((((...(((..(((((...)))))(((((((((.....)))))..)))))))..(.((((((....(((((.....)))))..)))))))......))).)))))))..... ( -47.80) >DroWil_CAF1 3723 120 + 1 UUGCCACCGCCCUCUAUGACUAUGAACCAAAGGCGGGUAGUAACCAACUGCAAUUGCGGACCGAUUGUCAGGUUCUGGUGAUUGGCAAAGAUGGCGAUAGUAAGGGCUGGUGGCGUGGCA ..(((((((((((.....(((((...(((...(((.(((((.....)))))...))).(.(((((..((((...))))..)))))).....)))..))))).))))).))))))...... ( -45.30) >DroAna_CAF1 4025 120 + 1 UUGCCACUGCCCUGUAUGAUUACGAACCAAAGGCUGGAAGCAAUCAACUGCAAUUGAGGACCGACUGCCAGGUCCUGGUCAUUGGCAAGGAUGGCGACAGCAAAGGCUGGUGGCGCGGCA ..(((((((((.((((....))))........((((...((.(((...(((...((((((((........)))))...)))...)))..))).))..))))...))).))))))...... ( -42.60) >DroPer_CAF1 2024 120 + 1 UAGCCACUGCCCUGUACGAUUACGAACCGAAGCCAGGGAGCAAUCAAUUGCAGCUGCGCACUGAUUGCCAGAUAUUGGUUAUUGGCAAAGAUGGGGACAGCAAGGGCUGGUGGCNNNNNN ..(((((((((((((...........((((((((((.(.(((((((..(((......))).)))))))).....)))))).))))......((....)))).))))).))))))...... ( -42.20) >consensus UAGCCACCGCCCUGUACGACUACGAACCAAAGGCGGGCAGCAAUCAACUGCAACUGCGCACCGACUGCCAGGUACUGGUCAUUGGCAAAGAUGGCGACAGCAAGGGCUGGUGGCGUGGCA ..(((((((((((.............((.......))..(((......)))...(((...((((..(((((...)))))..))))......((....)))))))))).))))))...... (-30.47 = -29.75 + -0.72)
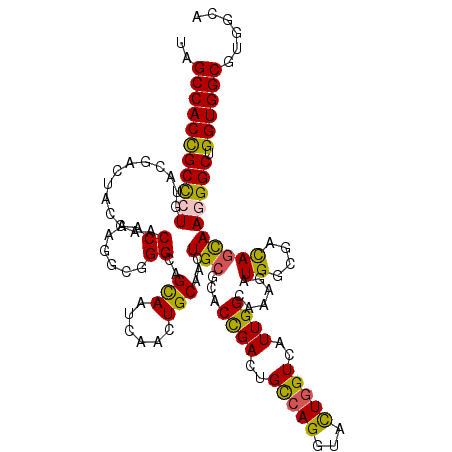

| Location | 19,105,257 – 19,105,377 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -41.31 |
| Consensus MFE | -28.03 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19105257 120 - 22224390 UGCCACGCCACCAGCCCUUGCUGUCCCCAUCCUUGCCAAUGACCAGCACCUGGCAGUCAGUGCGCAGCUGCAGCUGAUUGCUGCCCGCCUUUGGUUCGUAGUCGUAGAGAGCGGUGGCGA .....(((((((.((.(((((.............))..(((((..(.(((.(((.(.((((...((((....))))...)))).).)))...))).)...))))).))).))))))))). ( -43.22) >DroPse_CAF1 2015 120 - 1 UUCCUCGCCACCAGCCCUUGCUGUCCCCAUCUUUGCCAAUAACCAAUAUCUGGCAGUCGGUGCGCAGCUGCAAUUGAUUGCUCCCUGGCUUCGGUUCGUAAUCGUACAGGGCAGUGGCUA ......(((((..(((((.(((((...((((.(((((((((.....))).))))))..)))).))))).......((((((.....(((....))).))))))....))))).))))).. ( -41.00) >DroGri_CAF1 3824 120 - 1 UACCACGCCACCAGCCUUUGCUGUCGCCAUCCUUGCCAAUUACAAGUACCUGGCAAUCGGUACGCAGUUGCAGCUGAUUGCUGCCCGCCUUCGGAUCAAAGUCGUAUAGGGCGGUGGCGA .....(((((((.(((((.(((((.(((....((((((..((....))..))))))..)))..))))).(((((.....)))))(((....))).............)))))))))))). ( -45.40) >DroWil_CAF1 3723 120 - 1 UGCCACGCCACCAGCCCUUACUAUCGCCAUCUUUGCCAAUCACCAGAACCUGACAAUCGGUCCGCAAUUGCAGUUGGUUACUACCCGCCUUUGGUUCAUAGUCAUAGAGGGCGGUGGCAA ......((((((.(((((((((((.((((.....((((((.....(.(((........))).)((....)).)))))).............))))..)))))....)))))))))))).. ( -37.97) >DroAna_CAF1 4025 120 - 1 UGCCGCGCCACCAGCCUUUGCUGUCGCCAUCCUUGCCAAUGACCAGGACCUGGCAGUCGGUCCUCAAUUGCAGUUGAUUGCUUCCAGCCUUUGGUUCGUAAUCAUACAGGGCAGUGGCAA ((((((.....((((....))))..(((.....(((.(((....((((((........))))))..))))))(((((((((..((((...))))...))))))).))..))).)))))). ( -41.30) >DroPer_CAF1 2024 120 - 1 NNNNNNGCCACCAGCCCUUGCUGUCCCCAUCUUUGCCAAUAACCAAUAUCUGGCAAUCAGUGCGCAGCUGCAAUUGAUUGCUCCCUGGCUUCGGUUCGUAAUCGUACAGGGCAGUGGCUA ......(((((..(((((.(((((...(((..(((((((((.....))).))))))...))).))))).......((((((.....(((....))).))))))....))))).))))).. ( -39.00) >consensus UGCCACGCCACCAGCCCUUGCUGUCCCCAUCCUUGCCAAUAACCAGUACCUGGCAAUCGGUGCGCAGCUGCAGUUGAUUGCUCCCCGCCUUCGGUUCGUAAUCGUACAGGGCAGUGGCAA ......(((((..(((((.(((((.(((....((((((............))))))..)).).)))))......(((((((...(((....)))...)))))))...))))).))))).. (-28.03 = -27.87 + -0.16)

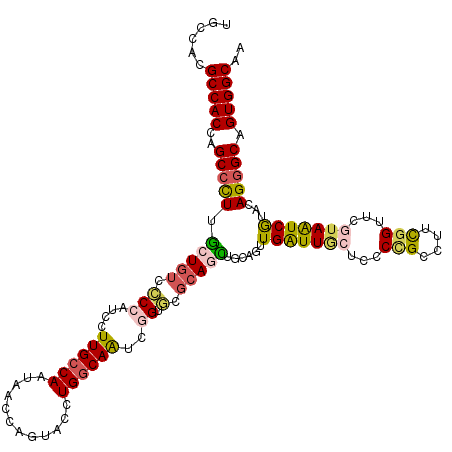

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:47 2006