
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,099,246 – 19,099,408 |
| Length | 162 |
| Max. P | 0.758344 |

| Location | 19,099,246 – 19,099,344 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -26.61 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
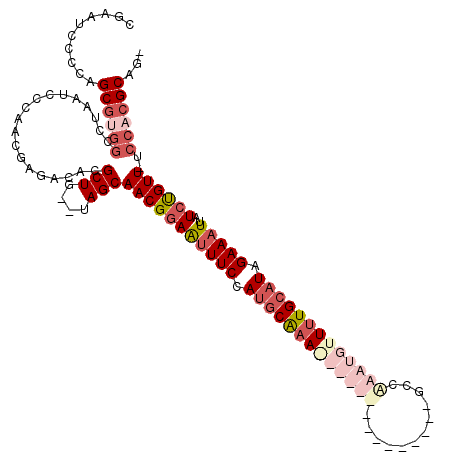
>X_DroMel_CAF1 19099246 98 - 22224390 ACAGCGCGUCUCGUUGGGAUUUGGCCACGCUUGGGAUGCGUUUUGAUAGUAGCAUCCCACC--CCGCUGGACACAAAGACAUUUAAUAUGUUGGACAGCA ...(((.((((.((.(((....(((...)))((((((((............))))))))))--).)).)))).)...(((((.....))))).....)). ( -31.00) >DroSec_CAF1 625 96 - 1 -CAGCGCGUCUCGUUGGGAUUAGGCCACGCUCGGGAUGCGUU-UGGUAGUGGCGUCCCACC--CCGCUGGACACAAAAACAUUUAAUAUGUUGGACAGCA -.(((((((((((.((((......)).))..)))))))))))-.((..((((....)))).--))((((..((.....((((.....))))))..)))). ( -33.10) >DroEre_CAF1 3102 90 - 1 -CAGCGUGUCUCGUUGGGAUUAGGAACUGCUGGGGAUUCGUU-U------AGGAUCCCACC--CCGCUGGACACAAAGACAUUUAAUAUGUUGGACAGCA -..((((((((.((.(((..(((......)))(((((((...-.------.))))))).))--).)).))))))...(((((.....))))).....)). ( -32.70) >DroYak_CAF1 2911 92 - 1 -CAGCGUUUCUCGUUGGGAUUAGGCCUCGCUGGGGAUUCGCU-C------AGGAUCCCACCCUCCGCUGGACACAAAGACAUUUAGUAUGUUGGACAGCA -(((((.....)))))(((...(((...))).(((((((...-.------.)))))))....)))((((..((((...((.....)).)).))..)))). ( -29.20) >consensus _CAGCGCGUCUCGUUGGGAUUAGGCCACGCUGGGGAUGCGUU_U______AGCAUCCCACC__CCGCUGGACACAAAGACAUUUAAUAUGUUGGACAGCA .(((((.....)))))((....((.....)).(((((((............))))))).....))((((..((.....((((.....))))))..)))). (-26.61 = -25.92 + -0.69)



| Location | 19,099,304 – 19,099,408 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.41 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -18.94 |
| Energy contribution | -23.63 |
| Covariance contribution | 4.69 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19099304 104 + 22224390 CGCAUCCCAAGCGUGGCCAAAUCCCAACGAGACGCGCUGUGUAGCAACGGAGUUUCCAUGCAAA---------------CCCAAAUGUUUUGCAUAGAAAUUAUCUGUU-UCCACGCAGC .((.((.....(((((.......)).))).)).))(((((((.(.(((((((((((.(((((((---------------.(.....).))))))).)))))..))))))-.).))))))) ( -30.90) >DroSec_CAF1 682 103 + 1 CGCAUCCCGAGCGUGGCCUAAUCCCAACGAGACGCGCUG--UAGCAACGGAGUUUCCAUGCAAA---------------GCCAAAUGCUAUGCAUCGAAAUUAUCCGUUUUCCACGCAGC ..........((((((.................((....--..))(((((((((((.(((((.(---------------((.....))).))))).)))))..))))))..))))))... ( -32.90) >DroEre_CAF1 3153 116 + 1 CGAAUCCCCAGCAGUUCCUAAUCCCAACGAGACACGCUG--UAGCAACGAAAUUUCCAUGCGAAGCAUUUAAAACAAAUGCCAAAUGUUUUGCAUAGAAUUUUUCUGUU-UCCACGCAG- ..........((.(((.........)))((((((.((..--..))...((((.(((.(((((((((((((............))))))))))))).)))..))))))))-))...))..- ( -25.20) >DroYak_CAF1 2964 116 + 1 CGAAUCCCCAGCGAGGCCUAAUCCCAACGAGAAACGCUG--UAGCAACGGAAUUUCCAAACGAAACAUUUAAAGCGAAUGCCGAAUGUUUUGCAUGGAAAUUGUCUGUU-UCCCCGCAG- ..........(((.((...................((..--..))(((((((((((((..((((((((((...((....)).))))))))))..)))))))..))))))-..)))))..- ( -29.60) >consensus CGAAUCCCCAGCGUGGCCUAAUCCCAACGAGACACGCUG__UAGCAACGGAAUUUCCAUGCAAA_______________GCCAAAUGUUUUGCAUAGAAAUUAUCUGUU_UCCACGCAG_ ..........((((((...................(((....)))(((((((((((.(((((((((((((............))))))))))))).)))))..))))))..))))))... (-18.94 = -23.63 + 4.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:44 2006