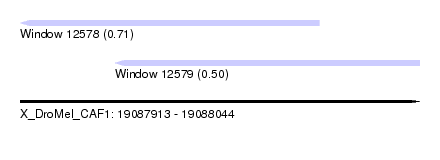
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,087,913 – 19,088,044 |
| Length | 131 |
| Max. P | 0.713049 |

| Location | 19,087,913 – 19,088,011 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -23.55 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
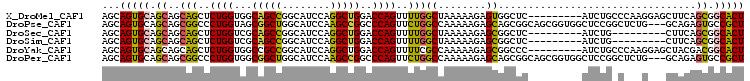
>X_DroMel_CAF1 19087913 98 - 22224390 AGCAGUGCAGCAGCAGCUCUGGUGGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGUGGCUC---------AUCUGCCCAAGGAGCUUCAGCGGCACU ...(((((.((.(.((((((...((((((((((......))))).(((.((((.......)))).)))...---------..)))))...)))))).).)).))))) ( -40.20) >DroPse_CAF1 4530 104 - 1 AGCAGUGCAGCAGCGGCCCUGGUAGCGGCUGGCAUCCAAGCCGGCCCAGUUCUGGCCAAAAAGAGCAGCGGCAGCGGUGGCUCCGGCUCUG---GCAGAGUGCCGCU (((((.((.(.(((.((((((...(((((((((......)))))))..(((((........))))).))..))).))).))).).)).))(---((.....)))))) ( -46.70) >DroSec_CAF1 4270 89 - 1 AGCAGUGCAGCAGCAGCUCUGGUCGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGCGGCUC---------AUCUG---------CUUCAGCGGCACU ...(((((.(((((....((((((.(((((........))))))))))).....))).....((((((...---------..)))---------)))..)).))))) ( -39.00) >DroSim_CAF1 4000 89 - 1 AGCAGUGCAGCAGCAGCUCUGGUCGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGCGGCUC---------AUCUG---------CUUCAGCGGCACU ...(((((.(((((....((((((.(((((........))))))))))).....))).....((((((...---------..)))---------)))..)).))))) ( -39.00) >DroYak_CAF1 4462 98 - 1 AGCAGUGCAGCAGCAGCUCUGGUGGCCGCCGGCAUCCAGGCUGGACCAGUUUUCGCCAAAAAGAGCGGCCC---------AUCUGCCCAAGGAGCUACGACGGCACU ...(((((.(..(.((((((((((((((((((...)).(((.(((......)))))).....).)))))).---------....)))...)))))).)..).))))) ( -35.20) >DroPer_CAF1 4519 104 - 1 AGCAGUGCAGCAGCGGCCCUGGUGGCGGCUGGCAUCCAAGCCGGCCCAGUUCUGGCCAAAAAGAGCAGCGGCAGCGGUGGCUCCGGCUCUG---GCAGAGUGCCGCU .((......))((((((((((.(((((((((((......)))))))((....))))))...(((((..(((.(((....))))))))))).---.))).).)))))) ( -49.20) >consensus AGCAGUGCAGCAGCAGCUCUGGUGGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCCAAAAAGAGCGGCUC_________AUCUG_C___G___CUACAGCGGCACU ...(((((.((..(((..((((...(((((........)))))..))))..)))((........)).................................)).))))) (-23.55 = -22.58 + -0.97)
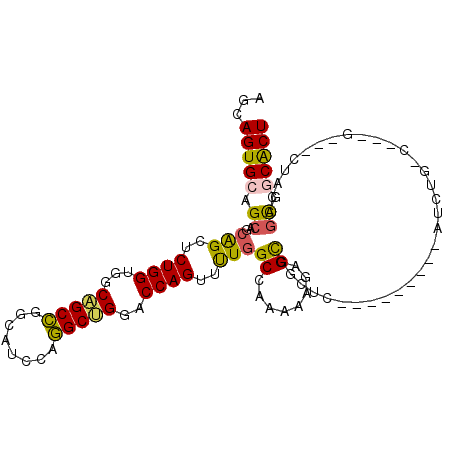

| Location | 19,087,944 – 19,088,044 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -35.94 |
| Energy contribution | -34.63 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19087944 100 - 22224390 -------GGAGGCGCAUCCCUCGCUCCCUGCGAGCUGUACAGCAGUGCAGCAGCAGCUCUGGUGGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGUG -------.((((......))))((((...((..(((((((....))))))).))(((((((((..(((((........))))).)))))....)))).....)))). ( -40.20) >DroPse_CAF1 4567 107 - 1 UGCAGGUGGUGGUGCGACCCUUGCACCCUGUGAGCUGUACAGCAGUGCAGCAGCGGCCCUGGUAGCGGCUGGCAUCCAAGCCGGCCCAGUUCUGGCCAAAAAGAGCA ..((((.(..(((((((...)))))))((((..(((((((....))))))).)))))))))...(.(((((((......)))))))).(((((........))))). ( -47.50) >DroSec_CAF1 4292 100 - 1 -------GGCGGCGCAUCCCUCGCUCCCUGCGAGCUGUACAGCAGUGCAGCAGCAGCUCUGGUCGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGCG -------((.((((.......)))).))(((..(((((((....))))))).)))(((((((((.(((((........)))))))))(((....)))....))))). ( -44.50) >DroSim_CAF1 4022 100 - 1 -------GGAGGCGCAUCCCUCGCUCCCUGCGAGCUGUACAGCAGUGCAGCAGCAGCUCUGGUCGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCUAAAAAGAGCG -------.((((......))))((((...((..(((((((....))))))).))((((((((((.(((((........)))))))))))....)))).....)))). ( -47.50) >DroYak_CAF1 4493 100 - 1 -------GGUGGCGCAUCCCUCGCUCCCUGCGAGCUGUACAGCAGUGCAGCAGCAGCUCUGGUGGCCGCCGGCAUCCAGGCUGGACCAGUUUUCGCCAAAAAGAGCG -------((..(((.......)))..))(((..(((((((....))))))).)))(((((..((((..(((((......)))))....(....)))))...))))). ( -41.60) >DroPer_CAF1 4556 107 - 1 UGGAGGUGGUGGUGCGACCCUAGCACCCUGUGAGCUGUACAGCAGUGCAGCAGCGGCCCUGGUGGCGGCUGGCAUCCAAGCCGGCCCAGUUCUGGCCAAAAAGAGCA .....(..(.(((((.......))))))..)..(((((((....))))))).((.((....)).))(((((((......)))))))..(((((........))))). ( -47.10) >consensus _______GGUGGCGCAUCCCUCGCUCCCUGCGAGCUGUACAGCAGUGCAGCAGCAGCUCUGGUGGCAGCCGGCAUCCAGGCUGGACCAGUUUUGGCCAAAAAGAGCG ...........((((.....((((.....))))(((((((....))))))).))((((((((...(((((........)))))..))))....)))).......)). (-35.94 = -34.63 + -1.30)


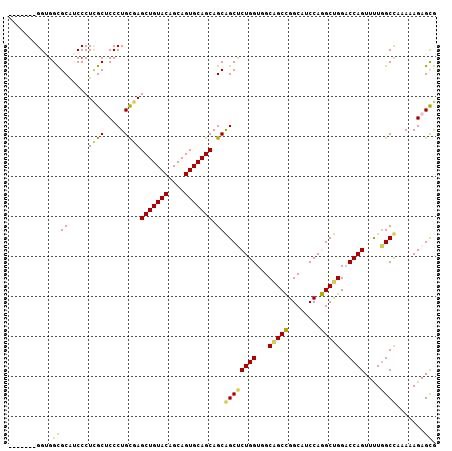
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:37 2006