
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,109,884 – 2,109,984 |
| Length | 100 |
| Max. P | 0.819596 |

| Location | 2,109,884 – 2,109,984 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2109884 100 + 22224390 UGUGCAUAUGGCACUAGCUAAGUGCAUGUCAGU---CCAACUUCAGCAAGGAGGAAGUGCCUUGCUAUCACUGGAGGUGCCUAAUGCACUUGACCUUUGGCAC---- .((((((..((((((.((.....)).......(---(((.....(((((((........))))))).....))))))))))..))))))..............---- ( -34.60) >DroSec_CAF1 3404 100 + 1 UGUGCAUAUGGCACUAGCUAAGUGCAUGUCAGU---CCAACUUCAGCAAGGAGGAAGUGCCUUGCUAUCACUGGAGGUGCCUAAUGCACUUGACCUUUGGCAC---- .((((((..((((((.((.....)).......(---(((.....(((((((........))))))).....))))))))))..))))))..............---- ( -34.60) >DroSim_CAF1 2048 100 + 1 UGUGCAUAUGGCACUAGCUAAGUGCAUGUCAGU---CCAACUUCAGCAAGGAGGAAGUGCCUUGCUAUCACUGGAGGUGCCUAAUGCACUUGACCUUUGGCAC---- .((((((..((((((.((.....)).......(---(((.....(((((((........))))))).....))))))))))..))))))..............---- ( -34.60) >DroEre_CAF1 3465 100 + 1 UGUGCAUAUAACACCAGCUGAGUGCAUGUCAGG---CCAACUUCUGCAAGGAGGAGCUGCAUUCAUAUCACUGGAGGUGGCUAGUGCACUUGACCUUUGGCAC---- .((((((....((((...((((((((.((....---((..(((....)))..)).))))))))))..((....))))))....))))))..............---- ( -27.50) >DroYak_CAF1 3327 101 + 1 UGUACACAUAGCACUGGCUAAGCGAAUGCCAGGUGCCAAACAUCUGCAAGUA------GCUUUCAUAUCACUGGAGGUGCCUAAUGCACUUGACCUUUGGCACUUGA ....(.(.((((....)))).).).....(((((((((((((..((((((..------(((((((......)))))))..))..))))..))...))))))))))). ( -31.60) >consensus UGUGCAUAUGGCACUAGCUAAGUGCAUGUCAGU___CCAACUUCAGCAAGGAGGAAGUGCCUUGCUAUCACUGGAGGUGCCUAAUGCACUUGACCUUUGGCAC____ .((((((.((((....)))).))))))(((((....(((.....(((((((........))))))).....)))((((((.....)))))).....)))))...... (-22.84 = -24.60 + 1.76)

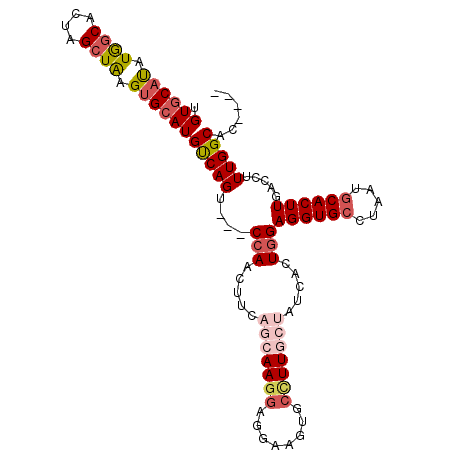

| Location | 2,109,884 – 2,109,984 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -32.71 |
| Consensus MFE | -23.08 |
| Energy contribution | -24.44 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2109884 100 - 22224390 ----GUGCCAAAGGUCAAGUGCAUUAGGCACCUCCAGUGAUAGCAAGGCACUUCCUCCUUGCUGAAGUUGG---ACUGACAUGCACUUAGCUAGUGCCAUAUGCACA ----((((.....((((.((((.....)))).((((((..((((((((........))))))))..)))))---).))))..(((((.....))))).....)))). ( -35.40) >DroSec_CAF1 3404 100 - 1 ----GUGCCAAAGGUCAAGUGCAUUAGGCACCUCCAGUGAUAGCAAGGCACUUCCUCCUUGCUGAAGUUGG---ACUGACAUGCACUUAGCUAGUGCCAUAUGCACA ----((((.....((((.((((.....)))).((((((..((((((((........))))))))..)))))---).))))..(((((.....))))).....)))). ( -35.40) >DroSim_CAF1 2048 100 - 1 ----GUGCCAAAGGUCAAGUGCAUUAGGCACCUCCAGUGAUAGCAAGGCACUUCCUCCUUGCUGAAGUUGG---ACUGACAUGCACUUAGCUAGUGCCAUAUGCACA ----((((.....((((.((((.....)))).((((((..((((((((........))))))))..)))))---).))))..(((((.....))))).....)))). ( -35.40) >DroEre_CAF1 3465 100 - 1 ----GUGCCAAAGGUCAAGUGCACUAGCCACCUCCAGUGAUAUGAAUGCAGCUCCUCCUUGCAGAAGUUGG---CCUGACAUGCACUCAGCUGGUGUUAUAUGCACA ----((((............(((((((((((.....)))...(((.((((((........))....(((..---...))).)))).))))))))))).....)))). ( -25.43) >DroYak_CAF1 3327 101 - 1 UCAAGUGCCAAAGGUCAAGUGCAUUAGGCACCUCCAGUGAUAUGAAAGC------UACUUGCAGAUGUUUGGCACCUGGCAUUCGCUUAGCCAGUGCUAUGUGUACA ....((((((((.(((..((((.....))))................((------.....)).))).)))))))).((((.........))))((((.....)))). ( -31.90) >consensus ____GUGCCAAAGGUCAAGUGCAUUAGGCACCUCCAGUGAUAGCAAGGCACUUCCUCCUUGCUGAAGUUGG___ACUGACAUGCACUUAGCUAGUGCCAUAUGCACA ..................((((((..(((((....((((.((((((((........))))))))..((..(....)..))...))))......)))))..)))))). (-23.08 = -24.44 + 1.36)

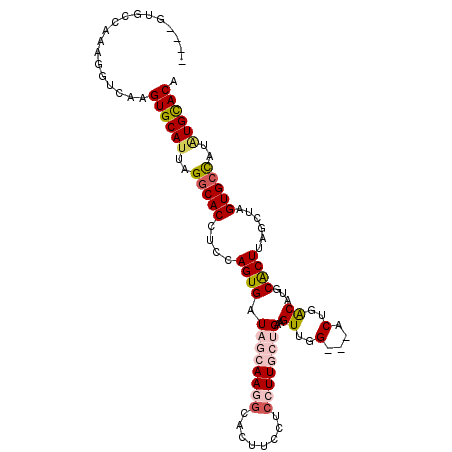
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:20 2006