
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,074,341 – 19,074,476 |
| Length | 135 |
| Max. P | 0.993917 |

| Location | 19,074,341 – 19,074,438 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
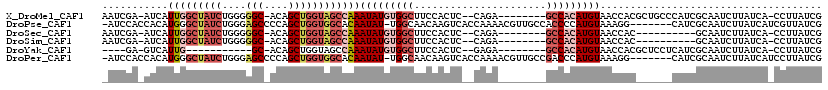
>X_DroMel_CAF1 19074341 97 + 22224390 AAGCAAAGCCCAGUGACUAUGGAUCCAUCGAUAAGGUGAUAAGAUUGCGAUGGGCAGCGUGGUUACAUGUGGCUCUGGAGUGGAAGCCACAUAUUUG ...((((.....(((((((((..(((((((.(((..(....)..))))))))))...)))))))))((((((((((.....)).)))))))).)))) ( -32.60) >DroSec_CAF1 25935 87 + 1 AAGCAAAGCCCAGUGACUAUGGAUCCAUCGAUAAGGUGAUAAGAUUGC----------GUGGUUACAUGUGGCUCUGGAGUGGAAGCCACAUAUUUG ...((((.....(((((((((((((((((.....))))....)))).)----------))))))))((((((((((.....)).)))))))).)))) ( -27.70) >DroSim_CAF1 27865 87 + 1 AAGCAAAGCCCAGUGACUAGGGAUCCAUCGAUAAGGUGAUAAGAUUGC----------GUGGUUACAUGUGGCUCUGGAGUGGAAGCCACAUAUUUG ...((((.....((((((((.((((((((.....))))....)))).)----------.)))))))((((((((((.....)).)))))))).)))) ( -26.50) >DroYak_CAF1 28417 97 + 1 AAGCAAAGCCCAGUGACUGUGGAUCCAUCGAUAAGGUGAUAAGAUUGCGAUGAGGAGCGUGGUUACAUGUGGCUCUCGAGUGGAAGCCACAUAUUUG ...((((.....(((((..((..(((((((.(((..(....)..)))))))).))..))..)))))((((((((..(....)..)))))))).)))) ( -29.40) >consensus AAGCAAAGCCCAGUGACUAUGGAUCCAUCGAUAAGGUGAUAAGAUUGC__________GUGGUUACAUGUGGCUCUGGAGUGGAAGCCACAUAUUUG ...((((.....(((((((((((((((((.....))))....))))...........)))))))))((((((((((.....)).)))))))).)))) (-21.46 = -22.02 + 0.56)



| Location | 19,074,341 – 19,074,438 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19074341 97 - 22224390 CAAAUAUGUGGCUUCCACUCCAGAGCCACAUGUAACCACGCUGCCCAUCGCAAUCUUAUCACCUUAUCGAUGGAUCCAUAGUCACUGGGCUUUGCUU ...(((((((((((........)))))))))))......((.(((((.........((((........))))(((.....)))..)))))...)).. ( -27.40) >DroSec_CAF1 25935 87 - 1 CAAAUAUGUGGCUUCCACUCCAGAGCCACAUGUAACCAC----------GCAAUCUUAUCACCUUAUCGAUGGAUCCAUAGUCACUGGGCUUUGCUU ...(((((((((((........)))))))))))......----------(((((((.(((........)))))).....((((....)))))))).. ( -21.90) >DroSim_CAF1 27865 87 - 1 CAAAUAUGUGGCUUCCACUCCAGAGCCACAUGUAACCAC----------GCAAUCUUAUCACCUUAUCGAUGGAUCCCUAGUCACUGGGCUUUGCUU ...(((((((((((........)))))))))))......----------(((((((.(((........))))))..(((((...)))))..)))).. ( -23.70) >DroYak_CAF1 28417 97 - 1 CAAAUAUGUGGCUUCCACUCGAGAGCCACAUGUAACCACGCUCCUCAUCGCAAUCUUAUCACCUUAUCGAUGGAUCCACAGUCACUGGGCUUUGCUU ...((((((((((..(....)..))))))))))......((((..(((((.................)))))(((.....)))...))))....... ( -23.73) >consensus CAAAUAUGUGGCUUCCACUCCAGAGCCACAUGUAACCAC__________GCAAUCUUAUCACCUUAUCGAUGGAUCCAUAGUCACUGGGCUUUGCUU ...(((((((((((........)))))))))))................(((((((.(((........)))))).....((((....)))))))).. (-21.77 = -21.77 + 0.00)



| Location | 19,074,369 – 19,074,476 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.40 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -13.69 |
| Energy contribution | -15.42 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
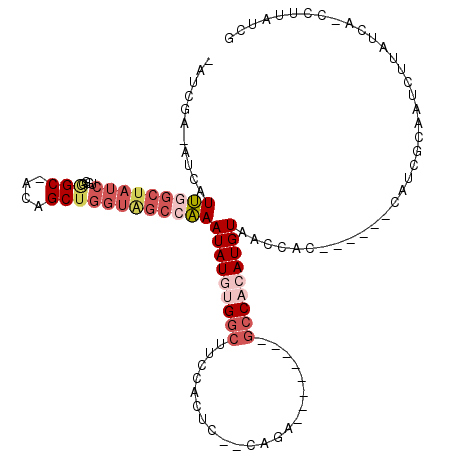
>X_DroMel_CAF1 19074369 107 - 22224390 AAUCGA-AUCAUUGGCUAUCUGGGGGC-ACAGCUGGUAGCCAAAUAUGUGGCUUCCACUC--CAGA--------GCCACAUGUAACCACGCUGCCCAUCGCAAUCUUAUCA-CCUUAUCG ...(((-....(((((((((((.....-.)))...))))))))(((((((((((......--..))--------)))))))))......((........))..........-.....))) ( -29.40) >DroPse_CAF1 29050 111 - 1 -AUCCACCACAUGGGCUAUCUGGGAGCCCCAGCUGGUGGCACAAUAU-UGGCAACAAGUCACCAAAACGUUGCCACCCCAUGUAAAGG-------CAUCGCAAUCUUAUCAUCGUUAUCG -..((...((((((((((((((((...)))))...))))).......-(((((((.............)))))))..))))))...))-------......................... ( -28.82) >DroSec_CAF1 25963 97 - 1 AAUCGA-AUCAUUGGCUAUCUGGGGGC-ACAGCUGGUAGCCAAAUAUGUGGCUUCCACUC--CAGA--------GCCACAUGUAACCAC----------GCAAUCUUAUCA-CCUUAUCG ...(((-....(((((((((((.....-.)))...))))))))(((((((((((......--..))--------)))))))))......----------............-.....))) ( -28.60) >DroSim_CAF1 27893 97 - 1 AAUCGA-AUCAUUGGCUAUCUGGGGGC-ACAGCUGGUAGCCAAAUAUGUGGCUUCCACUC--CAGA--------GCCACAUGUAACCAC----------GCAAUCUUAUCA-CCUUAUCG ...(((-....(((((((((((.....-.)))...))))))))(((((((((((......--..))--------)))))))))......----------............-.....))) ( -28.60) >DroYak_CAF1 28445 92 - 1 ----GA-GUCAUUG-----------GC-ACAGCUGGUAGCCAAAUAUGUGGCUUCCACUC--GAGA--------GCCACAUGUAACCACGCUCCUCAUCGCAAUCUUAUCA-CCUUAUCG ----((-((..(((-----------((-((.....)).)))))((((((((((..(....--)..)--------)))))))))......))))..................-........ ( -25.80) >DroPer_CAF1 30083 111 - 1 -AUCCACCACAUGGGCUAUCUGGGAGCCCCAGCUGGUGGCACAAUAU-UGGCAACAAGUCACCAAAACGUUGCCGACCCAUGUAAAGG-------CAUCGCAAUCUUAUCAUCCUUAUCG -..((...((((((((((((((((...)))))...)))))......(-(((((((.............)))))))).))))))...))-------......................... ( -29.52) >consensus _AUCGA_AUCAUUGGCUAUCUGGGGGC_ACAGCUGGUAGCCAAAUAUGUGGCUUCCACUC__CAGA________GCCACAUGUAACCAC______CAUCGCAAUCUUAUCA_CCUUAUCG ...........(((((((((....(((....))))))))))))(((((((((......................)))))))))..................................... (-13.69 = -15.42 + 1.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:32 2006