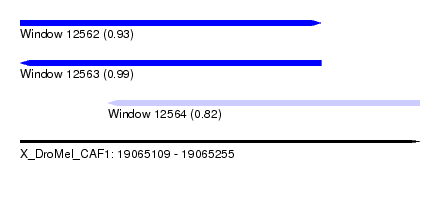
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,065,109 – 19,065,255 |
| Length | 146 |
| Max. P | 0.993737 |

| Location | 19,065,109 – 19,065,219 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.10 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19065109 110 + 22224390 GAUGCUGCGGUCGA--------GUGUGUGUGAAAAGGAAGUUGAGGGGAUUGAUUGGUCAGUAGCUG-UUGGCCAAUUGCCUUGGCUAAUUAUCAAUUUUGGACAACUGACCACCGACC ........((((.(--------((.(((.(((((..(((((..(((....(((((((((((......-))))))))))))))..))).....))..))))).))))))))))....... ( -32.40) >DroSec_CAF1 16825 101 + 1 -------CGGUCGA--------GUGUGUGUGAAAAGGAAGUUGAGGGGA---AUUGGCCAGUAGCUGGUUGGCCAAUUGUCUUGGCUAAUUAUCAAUUUUGGACAACUGACCACCGACC -------.((((.(--------((.(((.(((((..(((((..(((.((---.((((((((.......)))))))))).)))..))).....))..))))).))))))))))....... ( -33.80) >DroSim_CAF1 18808 101 + 1 -------CGGUCGA--------GUGUGUGUGAAAAGGAAGUUGAGGGGA---AUUGGCCAGUAGCUGGUUGGCCAAUUGUCUUGGCUAAUUAUCAAUUUCGGACAACUGACCACCGACC -------.((((.(--------((.(((.(((((..(((((..(((.((---.((((((((.......)))))))))).)))..))).....))..))))).))))))))))....... ( -36.60) >DroEre_CAF1 17238 105 + 1 AUUGUUGCGGUUGG----------GUGUGUGGAAAGGAAGUUGAGGGGA---GUUGGCCAGUAGCUGG-UGGCCAAUUGCAUGGGCUAAUUAUCAAUUUUGGACAACUGACCACCGACC ...((((.(((..(----------.(((.(..((..((((((.(..(.(---((((((((.(....).-))))))))).).).)))).....))..))..).))).)..)))..)))). ( -32.70) >DroYak_CAF1 18364 111 + 1 AAUGUUGCGGUCAGUUGUAUGUUUGUGUGCGAAAAGGAAGUUGAGGGGA---GUUGGCCAGUAGCUGG-UGGCCAAUUGCCUUGGCUAAUUAACAAUUUUGGACAACUGACCAAC---- ........((((((((((...((((....)))).....(((..(((.((---.(((((((.(....).-))))))))).)))..)))...............))))))))))...---- ( -41.60) >consensus __UG_UGCGGUCGA________GUGUGUGUGAAAAGGAAGUUGAGGGGA___AUUGGCCAGUAGCUGGUUGGCCAAUUGCCUUGGCUAAUUAUCAAUUUUGGACAACUGACCACCGACC ........(((((............(((.(((((..(((((..(((.(....(((((((((.......)))))))))).)))..))).....))..))))).)))..)))))....... (-26.74 = -26.10 + -0.64)


| Location | 19,065,109 – 19,065,219 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -20.23 |
| Energy contribution | -21.31 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19065109 110 - 22224390 GGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGGCAAUUGGCCAA-CAGCUACUGACCAAUCAAUCCCCUCAACUUCCUUUUCACACACAC--------UCGACCGCAGCAUC ((((((((((.(((((((.......))))))).(((((.....)))))...-..)))))))))).................................--------.............. ( -25.50) >DroSec_CAF1 16825 101 - 1 GGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGACAAUUGGCCAACCAGCUACUGGCCAAU---UCCCCUCAACUUCCUUUUCACACACAC--------UCGACCG------- (((((((((..(((((((.......)))))))..)))....(((((((((.........))))))))---)..........................--------)))))).------- ( -29.70) >DroSim_CAF1 18808 101 - 1 GGUCGGUGGUCAGUUGUCCGAAAUUGAUAAUUAGCCAAGACAAUUGGCCAACCAGCUACUGGCCAAU---UCCCCUCAACUUCCUUUUCACACACAC--------UCGACCG------- (((((((((..(((((((.......)))))))..)))....(((((((((.........))))))))---)..........................--------)))))).------- ( -29.70) >DroEre_CAF1 17238 105 - 1 GGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCCAUGCAAUUGGCCA-CCAGCUACUGGCCAAC---UCCCCUCAACUUCCUUUCCACACAC----------CCAACCGCAACAAU (((.(((((((((((......))))))).....((....))..(((((((-........))))))).---......................)))----------)..)))........ ( -24.50) >DroYak_CAF1 18364 111 - 1 ----GUUGGUCAGUUGUCCAAAAUUGUUAAUUAGCCAAGGCAAUUGGCCA-CCAGCUACUGGCCAAC---UCCCCUCAACUUCCUUUUCGCACACAAACAUACAACUGACCGCAACAUU ----((.((((((((((......((((......((....))..(((((((-........))))))).---.......................))))....))))))))))))...... ( -29.50) >consensus GGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGGCAAUUGGCCAACCAGCUACUGGCCAAU___UCCCCUCAACUUCCUUUUCACACACAC________UCGACCGCA_CA__ ((((((((((((((((((....................)))))))))))).((((...))))...........................................))))))........ (-20.23 = -21.31 + 1.08)



| Location | 19,065,141 – 19,065,255 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -25.75 |
| Energy contribution | -28.39 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19065141 114 - 22224390 UUUCUCCAGCUGGGUCGG----GCUGGUCAAUUGGGGCUGGGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGGCAAUUGGCCAA-CAGCUACUGACCAAUCAAUCCCCUCAA .....((((((......)----)))))......((((...((((((((((.(((((((.......))))))).(((((.....)))))...-..)))))))))).......)))).... ( -40.10) >DroSec_CAF1 16850 112 - 1 UUUCUCCUGGUGGGUCGG----GCUGGUCAAUUGGGGCUGGGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGACAAUUGGCCAACCAGCUACUGGCCAAU---UCCCCUCAA .......(((.(((...(----((..((.......((((((.....((((((((((((....................)))))))))))).))))))))..)))...---.))).))). ( -41.46) >DroSim_CAF1 18833 112 - 1 UUUCUCCAGGUGGGUCGG----GUUGGUCAAUUGGGGCUGGGUCGGUGGUCAGUUGUCCGAAAUUGAUAAUUAGCCAAGACAAUUGGCCAACCAGCUACUGGCCAAU---UCCCCUCAA ........(((.(((..(----((((((((((((...((.(((..((.(((((((......))))))).))..))).)).)))))))))))))....))).)))...---......... ( -44.00) >DroEre_CAF1 17268 115 - 1 UUUCUCCAGCUGGGUCGGUCGGGUUGCUCAAUUGGGGCUGGGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCCAUGCAAUUGGCCA-CCAGCUACUGGCCAAC---UCCCCUCAA ............(((((((..(((.((.((((((.(..(((((..((.(((((((......))))))).))..)))))).)))))))).)-))....)))))))...---......... ( -39.90) >DroYak_CAF1 18404 89 - 1 UUUCUCCUGGUGGGUCGG--------------------------GUUGGUCAGUUGUCCAAAAUUGUUAAUUAGCCAAGGCAAUUGGCCA-CCAGCUACUGGCCAAC---UCCCCUCAA .......(((.(((..((--------------------------((((((((((.((........(((((((.((....)))))))))..-...)).))))))))))---)))))))). ( -30.92) >consensus UUUCUCCAGGUGGGUCGG____GCUGGUCAAUUGGGGCUGGGUCGGUGGUCAGUUGUCCAAAAUUGAUAAUUAGCCAAGGCAAUUGGCCAACCAGCUACUGGCCAAU___UCCCCUCAA ..........((((..((......((((((.....((((((.....((((((((((((....................)))))))))))).))))))..))))))......)).)))). (-25.75 = -28.39 + 2.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:24 2006