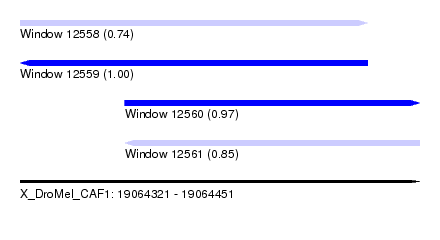
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 19,064,321 – 19,064,451 |
| Length | 130 |
| Max. P | 0.996784 |

| Location | 19,064,321 – 19,064,434 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -36.34 |
| Energy contribution | -36.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.739259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19064321 113 + 22224390 UUAGUGGCCCCCGCGGAUCUGCUUUUGUUUUGAUGCCCGCACCGUUGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCCCAGCUUCUGCUGCUCUACGAUGGGCUCU .....(((((..(((....)))....((((..((((((((.......))))))))....))))..(((.((((..(((.((((....))))..))))))).)))..))))).. ( -38.00) >DroSec_CAF1 16074 110 + 1 UUAGUGGCCCCCGCGGAUCUGCUUUUGUUUUGAUGCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCU ...((((...))))((.((.((....((((..((((((((.......))))))))....))))...)).)).))(((((..(..(((((....)))))..)---...))))). ( -41.00) >DroSim_CAF1 18049 110 + 1 UUAGUGGCCCCCGCGGAUCUGCUUUUGUUUUGAUGCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCU ...((((...))))((.((.((....((((..((((((((.......))))))))....))))...)).)).))(((((..(..(((((....)))))..)---...))))). ( -41.00) >DroEre_CAF1 16492 110 + 1 UUAGUGGCCCCCGCGGAUCUGCUUUUGUUUUGAUGCCCGCGCCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUGAGUCCCAGCUUCUGCUGCUCC---AUGGGCACU ..((((.((((.(((((((.((((..((((..((((((((.......))))))))....))))......)))).))((((.....)))).))))).)....---..))))))) ( -38.20) >DroYak_CAF1 17511 110 + 1 UUAGUAGCCCCCGCGGAUUUGCUUUUGUUUUGAUGCCCGCGCCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCU ..((.(((((.....(((((((((..((((..((((((((.......))))))))..............)))).)))).)))))(((((....)))))...---..))))))) ( -40.19) >consensus UUAGUGGCCCCCGCGGAUCUGCUUUUGUUUUGAUGCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC___AUGGGCUCU ......((((....(((((.((((..((((..((((((((.......))))))))....))))......)))).))........(((((....)))))))).....))))... (-36.34 = -36.42 + 0.08)

| Location | 19,064,321 – 19,064,434 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -34.82 |
| Energy contribution | -35.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19064321 113 - 22224390 AGAGCCCAUCGUAGAGCAGCAGAAGCUGGGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCAACGGUGCGGGCAUCAAAACAAAAGCAGAUCCGCGGGGGCCACUAA .(.((((.(((..((...((.((.((((((......))))))))..............(((((((((....).))))))))..........))...))..))))))))..... ( -40.10) >DroSec_CAF1 16074 110 - 1 AGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGCAUCAAAACAAAAGCAGAUCCGCGGGGGCCACUAA .((((....---(..(((((....)))))..)...))))(((((..............(((((((((....).))))))))..........((......))..)))))..... ( -41.50) >DroSim_CAF1 18049 110 - 1 AGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGCAUCAAAACAAAAGCAGAUCCGCGGGGGCCACUAA .((((....---(..(((((....)))))..)...))))(((((..............(((((((((....).))))))))..........((......))..)))))..... ( -41.50) >DroEre_CAF1 16492 110 - 1 AGUGCCCAU---GGAGCAGCAGAAGCUGGGACUCAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGCGCGGGCAUCAAAACAAAAGCAGAUCCGCGGGGGCCACUAA (((((((.(---(((...((.((.((((((......))))))))..............(((((((((....).))))))))..........))...))))...))).)))).. ( -40.00) >DroYak_CAF1 17511 110 - 1 AGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGCGCGGGCAUCAAAACAAAAGCAAAUCCGCGGGGGCUACUAA (((((((.(---((((((((....)))))......(((((......))..........(((((((((....).)))))))).........)))...))))...))))).)).. ( -39.70) >consensus AGAGCCCAU___GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGCAUCAAAACAAAAGCAGAUCCGCGGGGGCCACUAA .((((((.....)).(((((....)))))......))))(((((..............(((((((((....).))))))))..........((......))..)))))..... (-34.82 = -35.46 + 0.64)

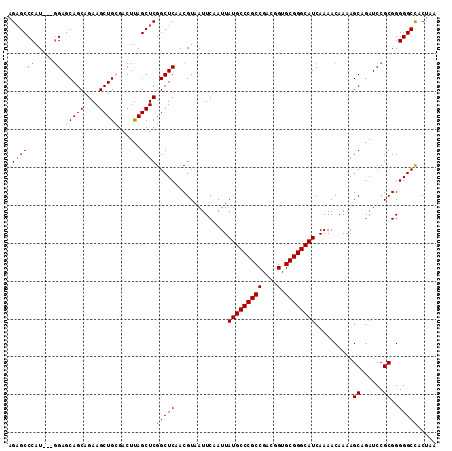
| Location | 19,064,355 – 19,064,451 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -33.10 |
| Energy contribution | -33.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19064355 96 + 22224390 GCCCGCACCGUUGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCCCAGCUUCUGCUGCUCUACGAUGGGCUCUACGUGGAAG--UAUC----CCAA ((((((.......)))))).........((((((.(((((.(....((...((((....))))..))....).))))).))))))...--....----.... ( -34.90) >DroSec_CAF1 16108 95 + 1 GCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCUACGUGGAAGUGUAUC----CCAA ((((((.......)))))).........((((((.(((((((......))(((((....)))))...---...))))).)))))).........----.... ( -37.20) >DroSim_CAF1 18083 95 + 1 GCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCUACGUGGAAGUGUAUC----CCAA ((((((.......)))))).........((((((.(((((((......))(((((....)))))...---...))))).)))))).........----.... ( -37.20) >DroEre_CAF1 16526 93 + 1 GCCCGCGCCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUGAGUCCCAGCUUCUGCUGCUCC---AUGGGCACUACGUGGAAG--UAUC----CCAA ((((((.......))))))....(((...(((....(((.((((((.....))))))..)))..(((---(((.......)))))).)--))..----.))) ( -32.30) >DroYak_CAF1 17545 97 + 1 GCCCGCGCCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC---AUGGGCUCUACAUGGAAG--UAUACCCCCGAA ((((((.......)))))).............((.(((((((......))(((((....)))))...---...))))).)).(((..(--....)..))).. ( -33.30) >consensus GCCCGCACCGUCGGCGGGCAUAAUUGAAUUACGUUGAGCCGAGCUAAGUCGCAGCUUCUGCUGCUCC___AUGGGCUCUACGUGGAAG__UAUC____CCAA ((((((.......)))))).........((((((.(((((.(........(((((....))))).......).))))).))))))................. (-33.10 = -33.90 + 0.80)

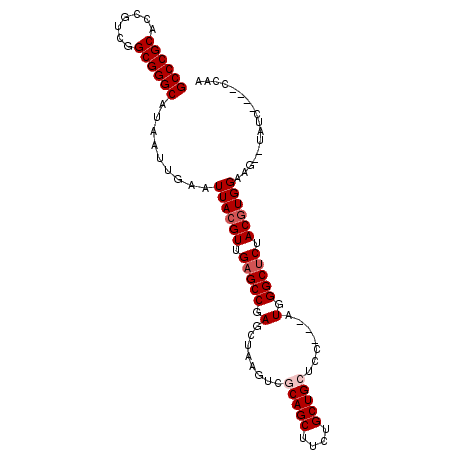

| Location | 19,064,355 – 19,064,451 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 91.77 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -29.55 |
| Energy contribution | -29.99 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 19064355 96 - 22224390 UUGG----GAUA--CUUCCACGUAGAGCCCAUCGUAGAGCAGCAGAAGCUGGGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCAACGGUGCGGGC ...(----((..--..)))((((.(((((....((.(((((((....))))...))).))..))))).))))...........(((((((....).)))))) ( -32.90) >DroSec_CAF1 16108 95 - 1 UUGG----GAUACACUUCCACGUAGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGC ...(----((......)))((((.(((((...---(..(((((....)))))..).......))))).))))...........(((((((....).)))))) ( -36.60) >DroSim_CAF1 18083 95 - 1 UUGG----GAUACACUUCCACGUAGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGC ...(----((......)))((((.(((((...---(..(((((....)))))..).......))))).))))...........(((((((....).)))))) ( -36.60) >DroEre_CAF1 16526 93 - 1 UUGG----GAUA--CUUCCACGUAGUGCCCAU---GGAGCAGCAGAAGCUGGGACUCAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGCGCGGGC .(((----(.((--((.......)))))))).---.((((....((.((((.....)))))).))))................(((((((....).)))))) ( -31.60) >DroYak_CAF1 17545 97 - 1 UUCGGGGGUAUA--CUUCCAUGUAGAGCCCAU---GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGCGCGGGC ...(((((....--)))))((((.(((((...---(..(((((....)))))..).......))))).))))...........(((((((....).)))))) ( -37.40) >consensus UUGG____GAUA__CUUCCACGUAGAGCCCAU___GGAGCAGCAGAAGCUGCGACUUAGCUCGGCUCAACGUAAUUCAAUUAUGCCCGCCGACGGUGCGGGC ...................((((.(((((.........(((((....)))))..........))))).))))...........(((((((....).)))))) (-29.55 = -29.99 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:44:21 2006